Tissue specificity of in vitro drug sensitivity
- PMID: 29016819
- PMCID: PMC6381764
- DOI: 10.1093/jamia/ocx062
Tissue specificity of in vitro drug sensitivity
Abstract
Objectives: We sought to investigate the tissue specificity of drug sensitivities in large-scale pharmacological studies and compare these associations to those found in drug clinical indications.
Materials and methods: We leveraged the curated cell line response data from PharmacoGx and applied an enrichment algorithm on drug sensitivity values' area under the drug dose-response curves (AUCs) with and without adjustment for general level of drug sensitivity.
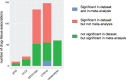
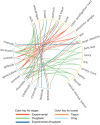
Results: We observed tissue specificity in 63% of tested drugs, with 8% of total interactions deemed significant (false discovery rate <0.05). By restricting the drug-tissue interactions to those with AUC > 0.2, we found that in 52% of interactions, the tissue was predictive of drug sensitivity (concordance index > 0.65). When compared with clinical indications, the observed overlap was weak (Matthew correlation coefficient, MCC = 0.0003, P > .10).
Discussion: While drugs exhibit significant tissue specificity in vitro, there is little overlap with clinical indications. This can be attributed to factors such as underlying biological differences between in vitro models and patient tumors, or the inability of tissue-specific drugs to bring additional benefits beyond gold standard treatments during clinical trials.
Conclusion: Our meta-analysis of pan-cancer drug screening datasets indicates that most tested drugs exhibit tissue-specific sensitivities in a large panel of cancer cell lines. However, the observed preclinical results do not translate to the clinical setting. Our results suggest that additional research into showing parallels between preclinical and clinical data is required to increase the translational potential of in vitro drug screening.
© The Author 2017. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Figures

Similar articles
-
Impact of between-tissue differences on pan-cancer predictions of drug sensitivity.PLoS Comput Biol. 2021 Feb 25;17(2):e1008720. doi: 10.1371/journal.pcbi.1008720. eCollection 2021 Feb. PLoS Comput Biol. 2021. PMID: 33630864 Free PMC article.
-
JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs.Bioorg Med Chem. 2012 Mar 15;20(6):1947-51. doi: 10.1016/j.bmc.2012.01.017. Epub 2012 Jan 21. Bioorg Med Chem. 2012. PMID: 22336246 Review.
-
Drug sensitivity in cancer cell lines is not tissue-specific.Mol Cancer. 2015 Feb 15;14:40. doi: 10.1186/s12943-015-0312-6. Mol Cancer. 2015. PMID: 25881072 Free PMC article.
-
Development of the Histoculture Drug Response Assay (HDRA).Methods Mol Biol. 2018;1760:39-48. doi: 10.1007/978-1-4939-7745-1_5. Methods Mol Biol. 2018. PMID: 29572792
-
Drug discovery strategies in the field of tumor energy metabolism: Limitations by metabolic flexibility and metabolic resistance to chemotherapy.Biochim Biophys Acta Bioenerg. 2017 Aug;1858(8):674-685. doi: 10.1016/j.bbabio.2017.02.005. Epub 2017 Feb 16. Biochim Biophys Acta Bioenerg. 2017. PMID: 28213330 Review.
Cited by
-
Discovering novel pharmacogenomic biomarkers by imputing drug response in cancer patients from large genomics studies.Genome Res. 2017 Oct;27(10):1743-1751. doi: 10.1101/gr.221077.117. Epub 2017 Aug 28. Genome Res. 2017. PMID: 28847918 Free PMC article.
-
Impact of between-tissue differences on pan-cancer predictions of drug sensitivity.PLoS Comput Biol. 2021 Feb 25;17(2):e1008720. doi: 10.1371/journal.pcbi.1008720. eCollection 2021 Feb. PLoS Comput Biol. 2021. PMID: 33630864 Free PMC article.
-
Anticancer drug synergy prediction in understudied tissues using transfer learning.J Am Med Inform Assoc. 2021 Jan 15;28(1):42-51. doi: 10.1093/jamia/ocaa212. J Am Med Inform Assoc. 2021. PMID: 33040150 Free PMC article.
-
Modeling Pathway Dynamics of the Skeletal Muscle Response to Intravenous Methylprednisolone (MPL) Administration in Rats: Dosing and Tissue Effects.Front Bioeng Biotechnol. 2020 Jul 14;8:759. doi: 10.3389/fbioe.2020.00759. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32760706 Free PMC article.
-
Tissue-guided LASSO for prediction of clinical drug response using preclinical samples.PLoS Comput Biol. 2020 Jan 22;16(1):e1007607. doi: 10.1371/journal.pcbi.1007607. eCollection 2020 Jan. PLoS Comput Biol. 2020. PMID: 31967990 Free PMC article.
References
-
- Wang B, Mezlini AM, Demir F, et al. . Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;113:333–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

