The HAND1 frameshift A126FS mutation does not cause hypoplastic left heart syndrome in mice
- PMID: 29016838
- PMCID: PMC5852545
- DOI: 10.1093/cvr/cvx166
The HAND1 frameshift A126FS mutation does not cause hypoplastic left heart syndrome in mice
Abstract
Aims: To test if a human Hand1 frame shift mutation identified in human samples is causative of hypoplastic left heart syndrome (HLHS).
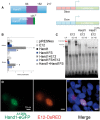
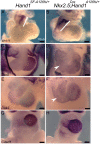
Methods and results: HLHS is a poorly understood single ventricle congenital heart defect that affects two to three infants in every 10 000 live births. The aetiologies of HLHS are largely unknown. The basic helix-loop-helix transcription factor HAND1 is required for normal heart development. Interrogation of HAND1 sequence from fixed HLHS tissues identified a somatic frame-shift mutation at Alanine 126 (NP_004812.1 p.Ala126Profs13X defined as Hand1A126fs). Hand1A126fs creates a truncated HAND1 protein that predictively functions as dominant negative. To determine if this mutation is causative of HLHS, we engineered a conditional Hand1A126fs mouse allele. Activation of this allele with Nkx2.5Cre results in E14.5 lethality accompanied by cardiac outflow tract and intraventricular septum abnormalities. Using αMHC-Cre or Mef2CAHF-Cre to activate Hand1A126fs results in reduced phenotype and limited viability. Left ventricles of Hand1A126FS mutant mice are not hypoplastic.
Conclusions: Somatically acquired Hand1A126FS mutation is not causative of HLHS. Hand1A126FS mutation does exhibit embryonic lethal cardiac defects that reflect a dominant negative function supporting the critical role of Hand1 in cardiogenesis.
Keywords: Cardiac development; Hand1; Hypoplastic left heart syndrome; Transcription; bHLH.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author 2017. For permissions, please email: journals.permissions@oup.com.
Figures





Similar articles
-
Directed differentiation of patient-specific induced pluripotent stem cells identifies the transcriptional repression and epigenetic modification of NKX2-5, HAND1, and NOTCH1 in hypoplastic left heart syndrome.PLoS One. 2014 Jul 22;9(7):e102796. doi: 10.1371/journal.pone.0102796. eCollection 2014. PLoS One. 2014. PMID: 25050861 Free PMC article.
-
A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts.Hum Mol Genet. 2008 May 15;17(10):1397-405. doi: 10.1093/hmg/ddn027. Epub 2008 Feb 14. Hum Mol Genet. 2008. PMID: 18276607
-
Somatic mutations in NKX2–5, GATA4, and HAND1 are not a common cause of tetralogy of Fallot or hypoplastic left heart.Am J Med Genet A. 2011 Oct;155A(10):2416-21. doi: 10.1002/ajmg.a.34187. Am J Med Genet A. 2011. PMID: 22043484
-
Gene replacement strategies to test the functional redundancy of basic helix-loop-helix transcription factor.Pediatr Cardiol. 2010 Apr;31(3):438-48. doi: 10.1007/s00246-010-9669-x. Epub 2010 Feb 14. Pediatr Cardiol. 2010. PMID: 20155416 Free PMC article. Review.
-
Partially Penetrant Cardiac Neural Crest Defects in Hand1 Phosphomutant Mice: Dimer Choice That Is Not So Critical.Pediatr Cardiol. 2019 Oct;40(7):1339-1344. doi: 10.1007/s00246-019-02162-8. Epub 2019 Jul 23. Pediatr Cardiol. 2019. PMID: 31338559 Free PMC article. Review.
Cited by
-
Conditional Mutation of Hand1 in the Mouse Placenta Disrupts Placental Vascular Development Resulting in Fetal Loss in Both Early and Late Pregnancy.Int J Mol Sci. 2021 Sep 2;22(17):9532. doi: 10.3390/ijms22179532. Int J Mol Sci. 2021. PMID: 34502440 Free PMC article.
-
Induced Pluripotent Stem Cell-Based Modeling of Single-Ventricle Congenital Heart Diseases.Curr Cardiol Rep. 2023 May;25(5):295-305. doi: 10.1007/s11886-023-01852-3. Epub 2023 Mar 17. Curr Cardiol Rep. 2023. PMID: 36930454 Free PMC article. Review.
-
Genetics of Congenital Heart Disease.Biomolecules. 2019 Dec 16;9(12):879. doi: 10.3390/biom9120879. Biomolecules. 2019. PMID: 31888141 Free PMC article. Review.
-
Identification and characterization of Hand2 upstream genomic enhancers active in developing stomach and limbs.Dev Dyn. 2024 Feb;253(2):215-232. doi: 10.1002/dvdy.646. Epub 2023 Aug 8. Dev Dyn. 2024. PMID: 37551791 Free PMC article.
-
Left-Sided Heart Defects and Laterality Disturbance in Hypoplastic Left Heart Syndrome.J Cardiovasc Dev Dis. 2023 Feb 24;10(3):99. doi: 10.3390/jcdd10030099. J Cardiovasc Dev Dis. 2023. PMID: 36975863 Free PMC article. Review.
References
-
- Tchervenkov CI, Jacobs ML, Tahta SA.. Congenital heart surgery nomenclature and database project: hypoplastic left heart syndrome. Ann Thorac Surg 2000;69:S170–S179. - PubMed
-
- Hinton RB, Martin LJ, Tabangin ME, Mazwi ML, Cripe LH, Benson W.. Hypoplastic left heart syndrome is heritable. J Am Coll Cardiol 2007;50:1590–1597. - PubMed
-
- Gordon BM, Rodriguez S, Lee M, Chang RK.. Decreasing number of deaths of infants with hypoplastic left heart syndrome. J Peds 2008;153:354–358. - PubMed
-
- Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, Obeid M, Bitar F.. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat 2006;27:293–294. - PubMed
-
- Liu X-Y, Wang J, Yang YQ, Zhang YY, Chen XZ, Zhang W, Wang XZ, Zheng J-H, Chen YH.. Novel NKX2-5 mutations in patients with familial atrial septal defects. Pediatr Cardiol 2011;32:193–201. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

