Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML
- PMID: 29017060
- PMCID: PMC7025434
- DOI: 10.1016/j.ccell.2017.09.004
Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML
Abstract
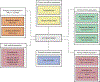
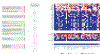
Chimeric antigen receptor (CAR) therapy targeting CD19 has yielded remarkable outcomes in patients with acute lymphoblastic leukemia. To identify potential CAR targets in acute myeloid leukemia (AML), we probed the AML surfaceome for overexpressed molecules with tolerable systemic expression. We integrated large transcriptomics and proteomics datasets from malignant and normal tissues, and developed an algorithm to identify potential targets expressed in leukemia stem cells, but not in normal CD34+CD38- hematopoietic cells, T cells, or vital tissues. As these investigations did not uncover candidate targets with a profile as favorable as CD19, we developed a generalizable combinatorial targeting strategy fulfilling stringent efficacy and safety criteria. Our findings indicate that several target pairings hold great promise for CAR therapy of AML.
Keywords: CAR T cell; acute myeloid leukemia (AML); algorithm; combinatorial strategies; high-throughput annotation; immunotherapy; leukemia; proteomics; surfaceome; target discovery.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures
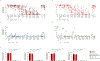


References
-
- (2016). ADCs Show Promise in Leukemias. Cancer Discov 6, 939. - PubMed
-
- Adam Mark RT, Afrasiabi Cyrus and Wu Chunlei (2014).
-
- Alvarez-Vallina L, and Hawkins RE (1996). Antigen-specific targeting of CD28-mediated T cell costimulation using chimeric single-chain antibody variable fragment-CD28 receptors. Eur J Immunol 26, 2304–2309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

