Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012-2014
- PMID: 29017452
- PMCID: PMC5634883
- DOI: 10.1186/s12879-017-2760-7
Whole genome sequencing for the molecular characterization of carbapenem-resistant Klebsiella pneumoniae strains isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012-2014
Abstract
Background: The emergence of carbapenem-resistant Klebsiella pneumoniae strains is threatening antimicrobial treatment.
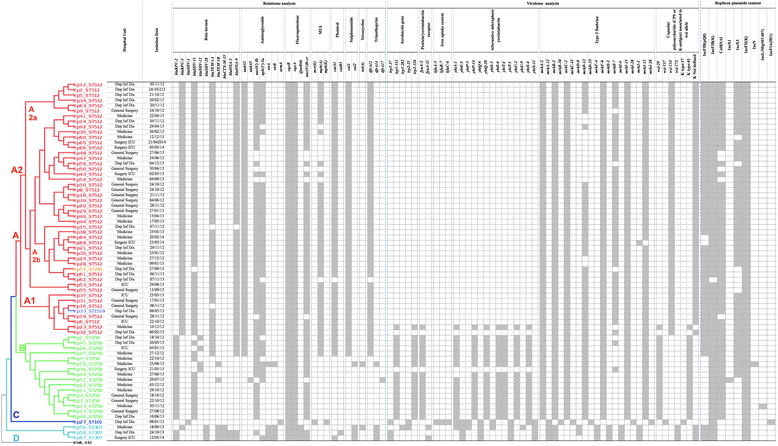
Methods: Sixty-eight carbapenemase-producing K. pneumoniae strains isolated at Luigi Sacco University Hospital-ASST Fatebenefratelli Sacco (Milan, Italy) between 2012 and 2014 were characterised microbiologically and molecularly. They were tested for drug susceptibility and carbapenemase phenotypes, investigated by means of repetitive extra-genic palindromic polymerase chain reaction (REP-PCR), and fully sequenced by means of next-generation sequencing for the in silico analysis of multi-locus sequence typing (MLST), their resistome, virulome and plasmid content, and their core single nucleotide polymorphism (SNP) genotypes.
Results: All of the samples were resistant to carbapenems, other β-lactams and ciprofloxacin; many were resistant to aminoglycosides and tigecycline; and seven were resistant to colistin. Resistome analysis revealed the presence of blaKPC genes and, less frequently blaSHV, blaTEM, blaCTX-M and blaOXA, which are related to resistance to carbapenem and other β-lactams. Other genes conferring resistance to aminoglycoside, fluoroquinolone, phenicol, sulphonamide, tetracycline, trimethoprim and macrolide-lincosamide-streptogramin were also detected. Genes related to AcrAB-TolC efflux pump-dependent and pump-independent tigecycline resistance mechanisms were investigated, but it was not possible to clearly correlate the genomic features with tigecycline resistance because of the presence of a common mutation in susceptible, intermediate and resistant strains. Concerning colistin resistance, the mgrB gene was disrupted by an IS5-like element, and the mobile mcr-1 and mcr-2 genes were not detected in two cases. The virulome profile revealed type-3 fimbriae and iron uptake system genes, which are important during the colonisation stage in the mammalian host environment. The in silico detected plasmid replicons were classified as IncFIB(pQil), IncFIB(K), ColRNAI, IncX1, IncX3, IncFII(K), IncN, IncL/M(pMU407) and IncFIA(HI1). REP-PCR showed five major clusters, and MLST revealed six different sequence types: 512, 258, 307, 1519, 745 and 101. Core SNP genotyping, which led to four clusters, correlated with the MLST data. Isolates of the same sequencing type often had common genetic traits, but the SNP analysis allowed greater strain tracking and discrimination than either the REP-PCR or MLST analysis.
Conclusion: Our findings support the importance of implementing bacterial genomics in clinical medicine in order to complement traditional methods and overcome their limited resolution.
Keywords: Bacteria epidemiology; Carbapenem-resistant Klebsiella pneumoniae; Whole-genome sequencing.
Conflict of interest statement
Ethics approval and consent to participate
All examined carbapenem-resistant
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy.BMC Infect Dis. 2019 Nov 4;19(1):928. doi: 10.1186/s12879-019-4565-3. BMC Infect Dis. 2019. PMID: 31684890 Free PMC article.
-
Genomic analysis of a pan-resistant Klebsiella pneumoniae sequence type 11 identified in Japan in 2016.Int J Antimicrob Agents. 2020 Apr;55(4):105854. doi: 10.1016/j.ijantimicag.2019.11.011. Epub 2019 Nov 23. Int J Antimicrob Agents. 2020. PMID: 31770626
-
An Update of the Evolving Epidemic of blaKPC Carrying Klebsiella pneumoniae in Sicily, Italy, 2014: Emergence of Multiple Non-ST258 Clones.PLoS One. 2015 Jul 15;10(7):e0132936. doi: 10.1371/journal.pone.0132936. eCollection 2015. PLoS One. 2015. PMID: 26177547 Free PMC article.
-
Prevalence of genes encoding carbapenem-resistance in Klebsiella pneumoniae recovered from clinical samples in Africa: systematic review and meta-analysis.BMC Infect Dis. 2025 Apr 18;25(1):556. doi: 10.1186/s12879-025-10959-7. BMC Infect Dis. 2025. PMID: 40251495 Free PMC article.
-
MgrB Mutations and Altered Cell Permeability in Colistin Resistance in Klebsiella pneumoniae.Cells. 2022 Sep 26;11(19):2995. doi: 10.3390/cells11192995. Cells. 2022. PMID: 36230959 Free PMC article. Review.
Cited by
-
Whole Genome Sequencing of Extended Spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae Isolated from Hospitalized Patients in KwaZulu-Natal, South Africa.Sci Rep. 2019 Apr 18;9(1):6266. doi: 10.1038/s41598-019-42672-2. Sci Rep. 2019. PMID: 31000772 Free PMC article.
-
Predicting Phenotypic Polymyxin Resistance in Klebsiella pneumoniae through Machine Learning Analysis of Genomic Data.mSystems. 2020 May 26;5(3):e00656-19. doi: 10.1128/mSystems.00656-19. mSystems. 2020. PMID: 32457240 Free PMC article.
-
Global prevalence of mutation in the mgrB gene among clinical isolates of colistin-resistant Klebsiella pneumoniae: a systematic review and meta-analysis.Front Microbiol. 2024 Jun 7;15:1386478. doi: 10.3389/fmicb.2024.1386478. eCollection 2024. Front Microbiol. 2024. PMID: 38912352 Free PMC article.
-
Identification and Antimicrobial Resistance in Klebsiella spp. Isolates from Turkeys in Poland between 2019 and 2022.Animals (Basel). 2022 Nov 15;12(22):3157. doi: 10.3390/ani12223157. Animals (Basel). 2022. PMID: 36428385 Free PMC article.
-
Computational design and characterization of a multiepitope vaccine against carbapenemase-producing Klebsiella pneumoniae strains, derived from antigens identified through reverse vaccinology.Comput Struct Biotechnol J. 2022 Aug 17;20:4446-4463. doi: 10.1016/j.csbj.2022.08.035. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36051872 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

