Lifestyle, gene gain and loss, and transcriptional remodeling cause divergence in the transcriptomes of Phytophthora infestans and Pythium ultimum during potato tuber colonization
- PMID: 29017458
- PMCID: PMC5635513
- DOI: 10.1186/s12864-017-4151-2
Lifestyle, gene gain and loss, and transcriptional remodeling cause divergence in the transcriptomes of Phytophthora infestans and Pythium ultimum during potato tuber colonization
Abstract
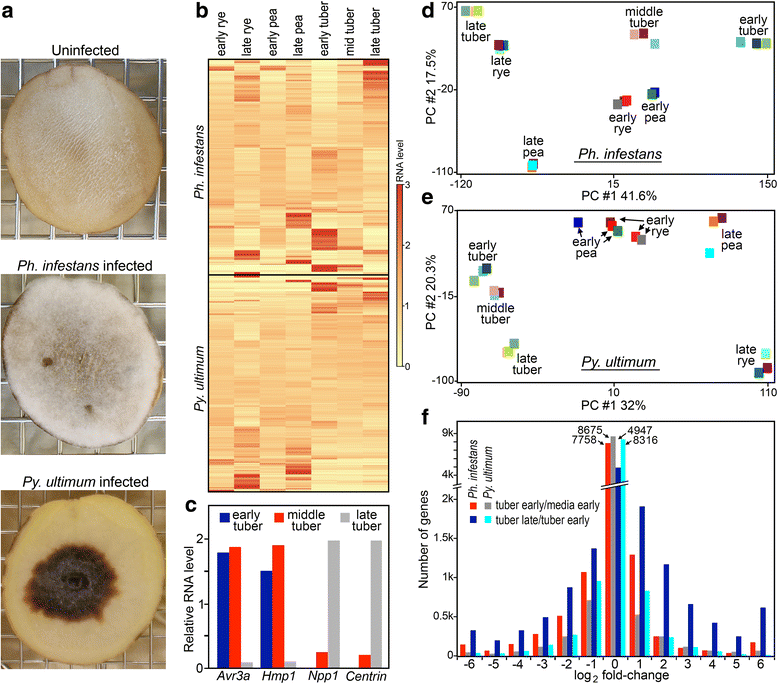
Background: How pathogen genomes evolve to support distinct lifestyles is not well-understood. The oomycete Phytophthora infestans, the potato blight agent, is a largely biotrophic pathogen that feeds from living host cells, which become necrotic only late in infection. The related oomycete Pythium ultimum grows saprophytically in soil and as a necrotroph in plants, causing massive tissue destruction. To learn what distinguishes their lifestyles, we compared their gene contents and expression patterns in media and a shared host, potato tuber.
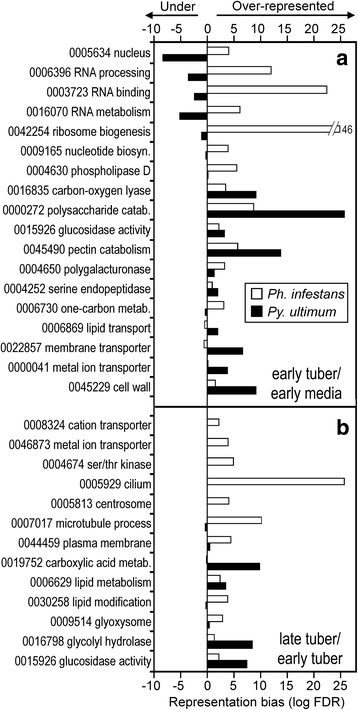
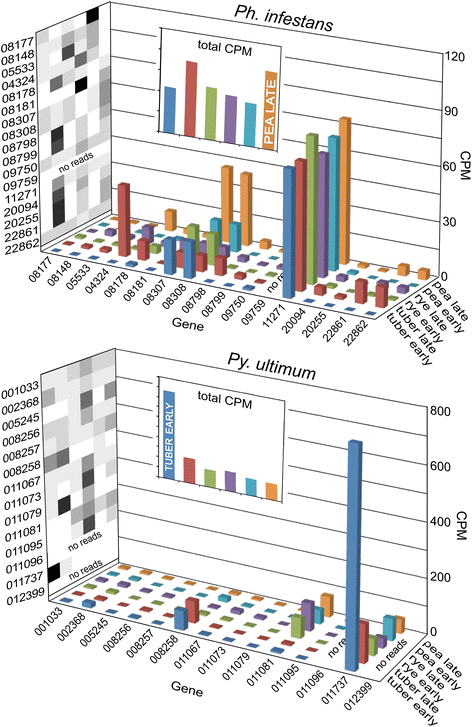
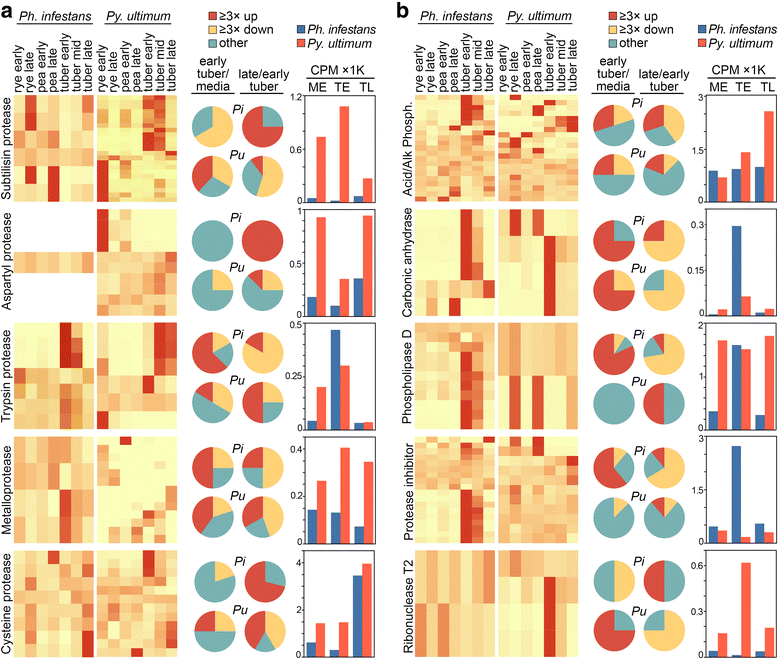
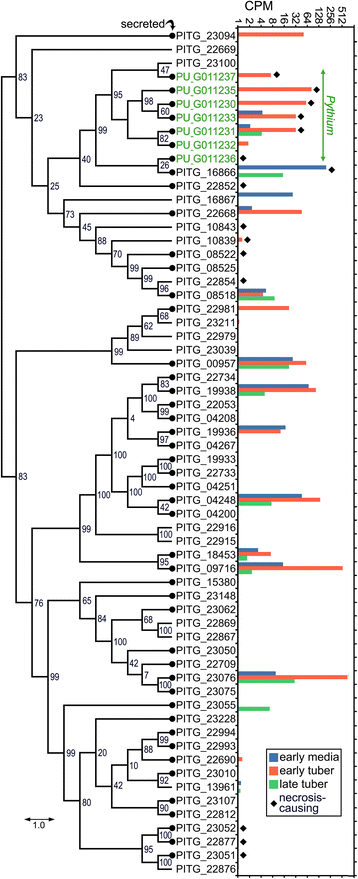
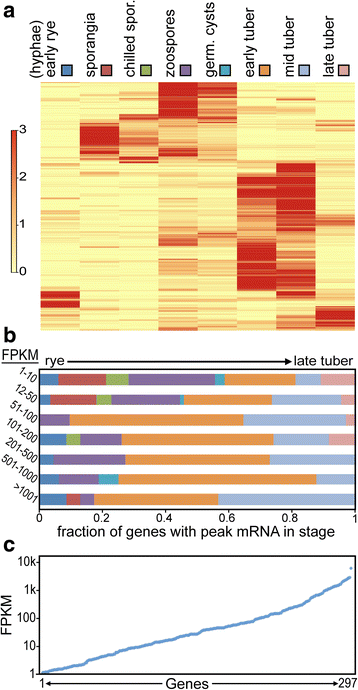
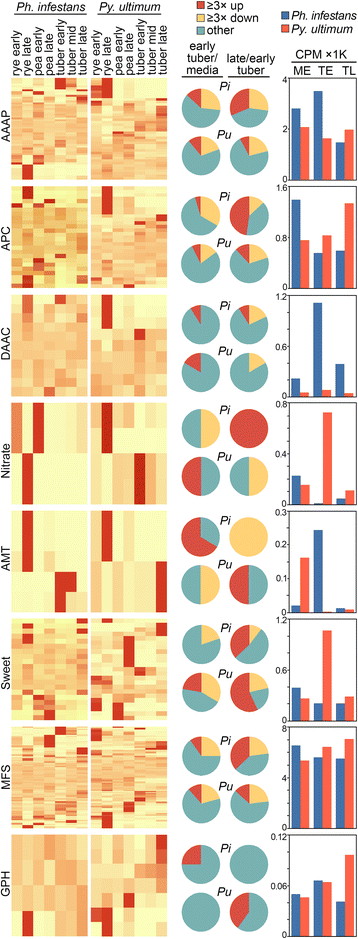
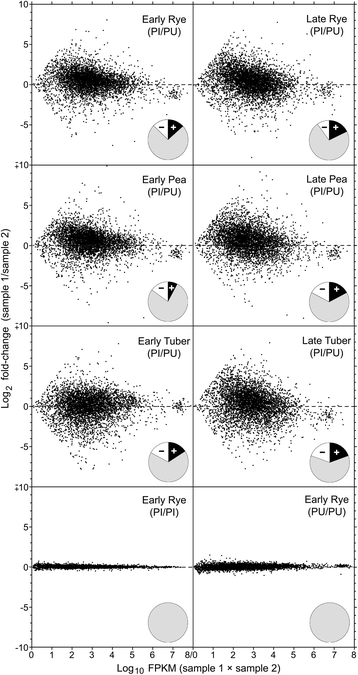
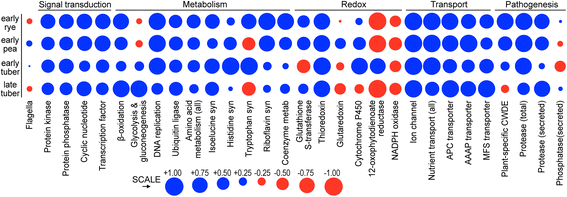
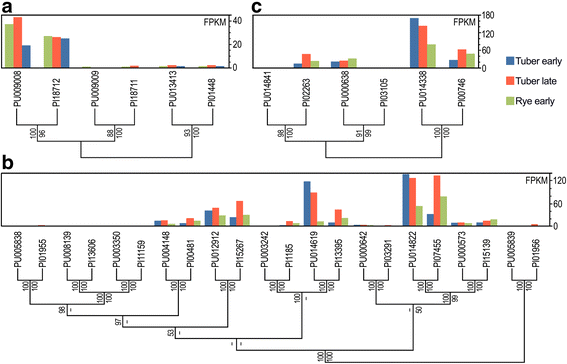
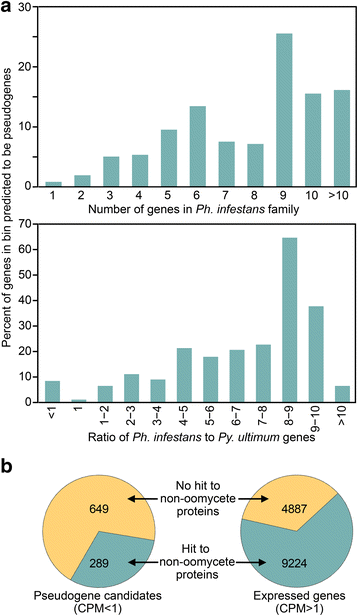
Results: Genes related to pathogenesis varied in temporal expression pattern, mRNA level, and family size between the species. A family's aggregate expression during infection was not proportional to size due to transcriptional remodeling and pseudogenization. Ph. infestans had more stage-specific genes, while Py. ultimum tended towards more constitutive expression. Ph. infestans expressed more genes encoding secreted cell wall-degrading enzymes, but other categories such as secreted proteases and ABC transporters had higher transcript levels in Py. ultimum. Species-specific genes were identified including new Pythium genes, perforins, which may disrupt plant membranes. Genome-wide ortholog analyses identified substantial diversified expression, which correlated with sequence divergence. Pseudogenization was associated with gene family expansion, especially in gene clusters.
Conclusion: This first large-scale analysis of transcriptional divergence within oomycetes revealed major shifts in genome composition and expression, including subfunctionalization within gene families. Biotrophy and necrotrophy seem determined by species-specific genes and the varied expression of shared pathogenicity factors, which may be useful targets for crop protection.
Keywords: Comparative genomics; Evolution; Gene family; Oomycete; Plant pathogen; RNA-seq; Regulatory subfunctionalization.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Lewis DH. Concepts in fungal nutrition and origin of biotrophy. Biol Rev Camb Philos Soc. 1973;48:261–278. doi: 10.1111/j.1469-185X.1973.tb00982.x. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

