The gut microbiome in atherosclerotic cardiovascular disease
- PMID: 29018189
- PMCID: PMC5635030
- DOI: 10.1038/s41467-017-00900-1
The gut microbiome in atherosclerotic cardiovascular disease
Abstract
The gut microbiota has been linked to cardiovascular diseases. However, the composition and functional capacity of the gut microbiome in relation to cardiovascular diseases have not been systematically examined. Here, we perform a metagenome-wide association study on stools from 218 individuals with atherosclerotic cardiovascular disease (ACVD) and 187 healthy controls. The ACVD gut microbiome deviates from the healthy status by increased abundance of Enterobacteriaceae and Streptococcus spp. and, functionally, in the potential for metabolism or transport of several molecules important for cardiovascular health. Although drug treatment represents a confounding factor, ACVD status, and not current drug use, is the major distinguishing feature in this cohort. We identify common themes by comparison with gut microbiome data associated with other cardiometabolic diseases (obesity and type 2 diabetes), with liver cirrhosis, and rheumatoid arthritis. Our data represent a comprehensive resource for further investigations on the role of the gut microbiome in promoting or preventing ACVD as well as other related diseases.The gut microbiota may play a role in cardiovascular diseases. Here, the authors perform a metagenome-wide association study on stools from individuals with atherosclerotic cardiovascular disease and healthy controls, identifying microbial strains and functions associated with the disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
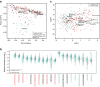



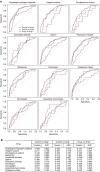



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

