PNLDC1 is essential for piRNA 3' end trimming and transposon silencing during spermatogenesis in mice
- PMID: 29018194
- PMCID: PMC5635004
- DOI: 10.1038/s41467-017-00854-4
PNLDC1 is essential for piRNA 3' end trimming and transposon silencing during spermatogenesis in mice
Abstract
Piwi-interacting RNAs are small regulatory RNAs with key roles in transposon silencing and regulation of gametogenesis. The production of mature piwi-interacting RNAs requires a critical step of trimming piwi-interacting RNA intermediates to achieve optimally sized piwi-interacting RNAs. The poly(A)-specific ribonuclease family deadenylase PNLDC1 is implicated in piwi-interacting RNA trimming in silkworms. The physiological function of PNLDC1 in mammals remains unknown. Using Pnldc1-deficient mice, here we show that PNLDC1 is required for piwi-interacting RNA biogenesis, transposon silencing, and spermatogenesis. Pnldc1 mutation in mice inhibits piwi-interacting RNA trimming and causes accumulation of untrimmed piwi-interacting RNA intermediates with 3' end extension, leading to severe reduction of mature piwi-interacting RNAs in the testis. Pnldc1 mutant mice exhibit disrupted LINE1 retrotransposon silencing and defect in spermiogenesis. Together, these results define PNLDC1 as a mammalian piwi-interacting RNA biogenesis factor that protects the germline genome and ensures normal sperm production in mice.piRNAs are regulatory RNAs that play a critical role in transposon silencing and gametogenesis. Here, the authors provide evidence that mammalian PNLDC1 is a regulator of piRNA biogenesis, transposon silencing and spermatogenesis, protecting the germline genome in mice.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
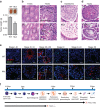
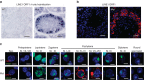

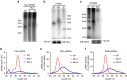
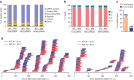
Comment in
-
Trimming it short: PNLDC1 is required for piRNA maturation during mouse spermatogenesis.EMBO Rep. 2018 Mar;19(3):e45824. doi: 10.15252/embr.201845824. Epub 2018 Feb 19. EMBO Rep. 2018. PMID: 29459487 Free PMC article.
Similar articles
-
PNLDC1 catalysis and postnatal germline function are required for piRNA trimming, LINE1 silencing, and spermatogenesis in mice.PLoS Genet. 2024 Sep 23;20(9):e1011429. doi: 10.1371/journal.pgen.1011429. eCollection 2024 Sep. PLoS Genet. 2024. PMID: 39312580 Free PMC article.
-
PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development.EMBO Rep. 2018 Mar;19(3):e44957. doi: 10.15252/embr.201744957. Epub 2018 Feb 14. EMBO Rep. 2018. PMID: 29444933 Free PMC article.
-
PNLDC1 catalysis and postnatal germline function are required for piRNA trimming, LINE1 silencing, and spermatogenesis in mice.bioRxiv [Preprint]. 2023 Dec 27:2023.12.26.573375. doi: 10.1101/2023.12.26.573375. bioRxiv. 2023. Update in: PLoS Genet. 2024 Sep 23;20(9):e1011429. doi: 10.1371/journal.pgen.1011429. PMID: 38234819 Free PMC article. Updated. Preprint.
-
PIWI-interacting RNAs (piRNAs) - a mouse testis perspective.Biochemistry (Mosc). 2013 Jun;78(6):592-602. doi: 10.1134/S0006297913060059. Biochemistry (Mosc). 2013. PMID: 23980886 Review.
-
The regulatory functions of piRNA/PIWI in spermatogenesis.Yi Chuan. 2017 Aug 20;39(8):683-691. doi: 10.16288/j.yczz.17-245. Yi Chuan. 2017. PMID: 28903896 Review.
Cited by
-
Sm complex assembly and 5' cap trimethylation promote selective processing of snRNAs by the 3' exonuclease TOE1.Proc Natl Acad Sci U S A. 2024 Jan 16;121(3):e2315259121. doi: 10.1073/pnas.2315259121. Epub 2024 Jan 9. Proc Natl Acad Sci U S A. 2024. PMID: 38194449 Free PMC article.
-
Co-option of the piRNA pathway to regulate neural crest specification.Sci Adv. 2022 Aug 12;8(32):eabn1441. doi: 10.1126/sciadv.abn1441. Epub 2022 Aug 10. Sci Adv. 2022. PMID: 35947657 Free PMC article.
-
Mini-Review Regarding the Applicability of Genome Editing Techniques Developed for Studying Infertility.Diagnostics (Basel). 2021 Feb 5;11(2):246. doi: 10.3390/diagnostics11020246. Diagnostics (Basel). 2021. PMID: 33562517 Free PMC article. Review.
-
An evolutionarily conserved stop codon enrichment at the 5' ends of mammalian piRNAs.Nat Commun. 2022 Apr 19;13(1):2118. doi: 10.1038/s41467-022-29787-3. Nat Commun. 2022. PMID: 35440552 Free PMC article.
-
MIWI N-terminal RG motif promotes efficient pachytene piRNA production and spermatogenesis independent of LINE1 transposon silencing.PLoS Genet. 2023 Nov 13;19(11):e1011031. doi: 10.1371/journal.pgen.1011031. eCollection 2023 Nov. PLoS Genet. 2023. PMID: 37956204 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

