Stress-induced plasticity of dynamic collagen networks
- PMID: 29018207
- PMCID: PMC5635002
- DOI: 10.1038/s41467-017-01011-7
Stress-induced plasticity of dynamic collagen networks
Abstract
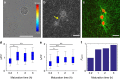
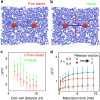
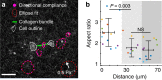
The structure and mechanics of tissues is constantly perturbed by endogenous forces originated from cells, and at the same time regulate many important cellular functions such as migration, differentiation, and growth. Here we show that 3D collagen gels, major components of connective tissues and extracellular matrix (ECM), are significantly and irreversibly remodeled by cellular traction forces, as well as by macroscopic strains. To understand this ECM plasticity, we develop a computational model that takes into account the sliding and merging of ECM fibers. We have confirmed the model predictions with experiment. Our results suggest the profound impacts of cellular traction forces on their host ECM during development and cancer progression, and suggest indirect mechanical channels of cell-cell communications in 3D fibrous matrices.The structure and mechanics of tissues is constantly perturbed by endogenous forces originated from cells. Here the authors show that 3D collagen gels, major components of connective tissues and extracellular matrix, are significantly and irreversibly remodelled by cellular traction forces and by macroscopic strains.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Effects of extracellular fiber architecture on cell membrane shear stress in a 3D fibrous matrix.J Biomech. 2007;40(7):1484-92. doi: 10.1016/j.jbiomech.2006.06.023. Epub 2006 Sep 20. J Biomech. 2007. PMID: 16987520
-
Computational model for cell migration in three-dimensional matrices.Biophys J. 2005 Aug;89(2):1389-97. doi: 10.1529/biophysj.105.060723. Epub 2005 May 20. Biophys J. 2005. PMID: 15908579 Free PMC article.
-
Toward single cell traction microscopy within 3D collagen matrices.Exp Cell Res. 2013 Oct 1;319(16):2396-408. doi: 10.1016/j.yexcr.2013.06.009. Epub 2013 Jun 25. Exp Cell Res. 2013. PMID: 23806281 Free PMC article. Review.
-
Tissue engineering science: consequences of cell traction force.Cytotechnology. 1992;10(3):225-50. doi: 10.1007/BF00146673. Cytotechnology. 1992. PMID: 1369238
-
Mechanical induction of gene expression in connective tissue cells.Methods Cell Biol. 2010;98:178-205. doi: 10.1016/S0091-679X(10)98008-4. Methods Cell Biol. 2010. PMID: 20816235 Review.
Cited by
-
Effects of extracellular matrix viscoelasticity on cellular behaviour.Nature. 2020 Aug;584(7822):535-546. doi: 10.1038/s41586-020-2612-2. Epub 2020 Aug 26. Nature. 2020. PMID: 32848221 Free PMC article. Review.
-
Compressive instabilities enable cell-induced extreme densification patterns in the fibrous extracellular matrix: Discrete model predictions.PLoS Comput Biol. 2024 Jul 1;20(7):e1012238. doi: 10.1371/journal.pcbi.1012238. eCollection 2024 Jul. PLoS Comput Biol. 2024. PMID: 38950077 Free PMC article.
-
Fibrin prestress due to platelet aggregation and contraction increases clot stiffness.Biophys Rep (N Y). 2021 Sep 14;1(2):100022. doi: 10.1016/j.bpr.2021.100022. eCollection 2021 Dec 8. Biophys Rep (N Y). 2021. PMID: 36425457 Free PMC article.
-
Selection of natural biomaterials for micro-tissue and organ-on-chip models.J Biomed Mater Res A. 2022 May;110(5):1147-1165. doi: 10.1002/jbm.a.37353. Epub 2022 Jan 31. J Biomed Mater Res A. 2022. PMID: 35102687 Free PMC article. Review.
-
Effect of matrix heterogeneity on cell mechanosensing.Soft Matter. 2021 Nov 24;17(45):10263-10273. doi: 10.1039/d1sm00312g. Soft Matter. 2021. PMID: 34125129 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

