Codon optimality, bias and usage in translation and mRNA decay
- PMID: 29018283
- PMCID: PMC6594389
- DOI: 10.1038/nrm.2017.91
Codon optimality, bias and usage in translation and mRNA decay
Abstract
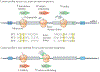
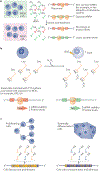
The advent of ribosome profiling and other tools to probe mRNA translation has revealed that codon bias - the uneven use of synonymous codons in the transcriptome - serves as a secondary genetic code: a code that guides the efficiency of protein production, the fidelity of translation and the metabolism of mRNAs. Recent advancements in our understanding of mRNA decay have revealed a tight coupling between ribosome dynamics and the stability of mRNA transcripts; this coupling integrates codon bias into the concept of codon optimality, or the effects that specific codons and tRNA concentrations have on the efficiency and fidelity of the translation machinery. In this Review, we first discuss the evidence for codon-dependent effects on translation, beginning with the basic mechanisms through which translation perturbation can affect translation efficiency, protein folding and transcript stability. We then discuss how codon effects are leveraged by the cell to tailor the proteome to maintain homeostasis, execute specific gene expression programmes of growth or differentiation and optimize the efficiency of protein production.
Figures


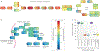
References
-
- Ikemura T Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151, 389–409 (1981). - PubMed
-
- Duret L tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16, 287–289 (2000). - PubMed
-
- Moriyama EN & Powell JR Codon usage bias and tRNA abundance in Drosophila. J. Mol. Evol. 45, 514–523 (1997). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

