Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding
- PMID: 29019322
- PMCID: PMC5665648
- DOI: 10.7554/eLife.31652
Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding
Abstract
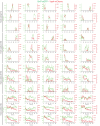
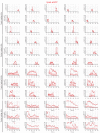
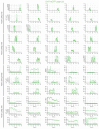
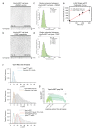
The ESCRT machinery mediates reverse membrane scission. By quantitative fluorescence lattice light-sheet microscopy, we have shown that ESCRT-III subunits polymerize rapidly on yeast endosomes, together with the recruitment of at least two Vps4 hexamers. During their 3-45 s lifetimes, the ESCRT-III assemblies accumulated 75-200 Snf7 and 15-50 Vps24 molecules. Productive budding events required at least two additional Vps4 hexamers. Membrane budding was associated with continuous, stochastic exchange of Vps4 and ESCRT-III components, rather than steady growth of fixed assemblies, and depended on Vps4 ATPase activity. An all-or-none step led to final release of ESCRT-III and Vps4. Tomographic electron microscopy demonstrated that acute disruption of Vps4 recruitment stalled membrane budding. We propose a model in which multiple Vps4 hexamers (four or more) draw together several ESCRT-III filaments. This process induces cargo crowding and inward membrane buckling, followed by constriction of the nascent bud neck and ultimately ILV generation by vesicle fission.
Keywords: 3D tracking; ESCRT; S. cerevisiae; cell biology; electron tomography; endosomes; high pressure freeze substitution; lattice light sheet microscopy; membrane budding.
Conflict of interest statement
No competing interests declared.
Figures











References
-
- Aguet F, Upadhyayula S, Gaudin R, Chou YY, Cocucci E, He K, Chen BC, Mosaliganti K, Pasham M, Skillern W, Legant WR, Liu TL, Findlay G, Marino E, Danuser G, Megason S, Betzig E, Kirchhausen T. Membrane dynamics of dividing cells imaged by lattice light-sheet microscopy. Molecular Biology of the Cell. 2016;27:3418–3435. doi: 10.1091/mbc.E16-03-0164. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

