Radically truncated MeCP2 rescues Rett syndrome-like neurological defects
- PMID: 29019980
- PMCID: PMC5884422
- DOI: 10.1038/nature24058
Radically truncated MeCP2 rescues Rett syndrome-like neurological defects
Abstract
Heterozygous mutations in the X-linked MECP2 gene cause the neurological disorder Rett syndrome. The methyl-CpG-binding protein 2 (MeCP2) protein is an epigenetic reader whose binding to chromatin primarily depends on 5-methylcytosine. Functionally, MeCP2 has been implicated in several cellular processes on the basis of its reported interaction with more than 40 binding partners, including transcriptional co-repressors (for example, the NCoR/SMRT complex), transcriptional activators, RNA, chromatin remodellers, microRNA-processing proteins and splicing factors. Accordingly, MeCP2 has been cast as a multi-functional hub that integrates diverse processes that are essential in mature neurons. At odds with the concept of broad functionality, missense mutations that cause Rett syndrome are concentrated in two discrete clusters coinciding with interaction sites for partner macromolecules: the methyl-CpG binding domain and the NCoR/SMRT interaction domain. Here we test the hypothesis that the single dominant function of MeCP2 is to physically connect DNA with the NCoR/SMRT complex, by removing almost all amino-acid sequences except the methyl-CpG binding and NCoR/SMRT interaction domains. We find that mice expressing truncated MeCP2 lacking both the N- and C-terminal regions (approximately half of the native protein) are phenotypically near-normal; and those expressing a minimal MeCP2 additionally lacking a central domain survive for over one year with only mild symptoms. This minimal protein is able to prevent or reverse neurological symptoms when introduced into MeCP2-deficient mice by genetic activation or virus-mediated delivery to the brain. Thus, despite evolutionary conservation of the entire MeCP2 protein sequence, the DNA and co-repressor binding domains alone are sufficient to avoid Rett syndrome-like defects and may therefore have therapeutic utility.
Conflict of interest statement
Conflict of interest statement: A.B. is a member of the Board of ArRETT, a company based in the United States with the goal of developing therapies for Rett syndrome.
Figures

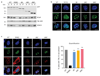

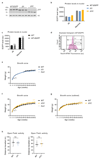







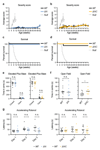
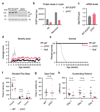

Comment in
-
Neurobiology: Domains to the rescue for Rett syndrome.Nature. 2017 Oct 19;550(7676):343-344. doi: 10.1038/nature24151. Epub 2017 Oct 11. Nature. 2017. PMID: 29019977 No abstract available.
References
-
- Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. - PubMed
-
- Lewis JD, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–14. - PubMed
-
- Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat Rev Genet. 2015;16:261–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

