The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3'-end processing
- PMID: 29020104
- PMCID: PMC5636114
- DOI: 10.1371/journal.pone.0186034
The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3'-end processing
Abstract
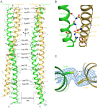
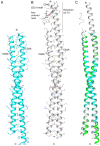
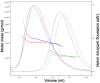
Unlike canonical pre-mRNAs, animal replication-dependent histone pre-mRNAs lack introns and are processed at the 3'-end by a mechanism distinct from cleavage and polyadenylation. They have a 3' stem loop and histone downstream element (HDE) that are recognized by stem-loop binding protein (SLBP) and U7 snRNP, respectively. The N-terminal domain (NTD) of Lsm11, a component of U7 snRNP, interacts with FLASH NTD and these two proteins recruit the histone cleavage complex containing the CPSF-73 endonuclease for the cleavage reaction. Here, we determined crystal structures of FLASH NTD and found that it forms a coiled-coil dimer. Using solution light scattering, we characterized the stoichiometry of the FLASH NTD-Lsm11 NTD complex and found that it is a 2:1 heterotrimer, which is supported by observations from analytical ultracentrifugation and crosslinking.
Conflict of interest statement
Figures



Similar articles
-
U7 deciphered: the mechanism that forms the unusual 3' end of metazoan replication-dependent histone mRNAs.Biochem Soc Trans. 2021 Nov 1;49(5):2229-2240. doi: 10.1042/BST20210323. Biochem Soc Trans. 2021. PMID: 34351387 Free PMC article. Review.
-
U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein.RNA. 2017 Jun;23(6):938-951. doi: 10.1261/rna.060806.117. Epub 2017 Mar 13. RNA. 2017. PMID: 28289156 Free PMC article.
-
3'-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors.RNA. 2013 Dec;19(12):1726-44. doi: 10.1261/rna.040360.113. Epub 2013 Oct 21. RNA. 2013. PMID: 24145821 Free PMC article.
-
A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3'-end processing.Mol Cell Biol. 2013 Jan;33(1):28-37. doi: 10.1128/MCB.00653-12. Epub 2012 Oct 15. Mol Cell Biol. 2013. PMID: 23071092 Free PMC article.
-
Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body.RNA Biol. 2017 Jun 3;14(6):726-738. doi: 10.1080/15476286.2016.1265198. Epub 2017 Jan 6. RNA Biol. 2017. PMID: 28059623 Free PMC article. Review.
Cited by
-
A real-time fluorescence assay for CPSF73, the nuclease for pre-mRNA 3'-end processing.RNA. 2021 Oct;27(10):1148-1154. doi: 10.1261/rna.078764.121. Epub 2021 Jul 6. RNA. 2021. PMID: 34230059 Free PMC article.
-
Spatiotemporal higher-order chromatin landscape of human histone gene clusters at histone locus bodies during the cell cycle in breast cancer progression.Gene. 2023 Jul 1;872:147441. doi: 10.1016/j.gene.2023.147441. Epub 2023 Apr 23. Gene. 2023. PMID: 37094694 Free PMC article.
-
Studies with recombinant U7 snRNP demonstrate that CPSF73 is both an endonuclease and a 5'-3' exonuclease.RNA. 2020 Oct;26(10):1345-1359. doi: 10.1261/rna.076273.120. Epub 2020 Jun 17. RNA. 2020. PMID: 32554553 Free PMC article.
-
U7 deciphered: the mechanism that forms the unusual 3' end of metazoan replication-dependent histone mRNAs.Biochem Soc Trans. 2021 Nov 1;49(5):2229-2240. doi: 10.1042/BST20210323. Biochem Soc Trans. 2021. PMID: 34351387 Free PMC article. Review.
-
Reconstitution and biochemical assays of an active human histone pre-mRNA 3'-end processing machinery.Methods Enzymol. 2021;655:291-324. doi: 10.1016/bs.mie.2021.03.021. Epub 2021 May 3. Methods Enzymol. 2021. PMID: 34183127 Free PMC article.
References
-
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–54. doi: 10.1038/nrg2438 - DOI - PMC - PubMed
-
- Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–80. doi: 10.1016/j.ceb.2005.04.010 - DOI - PubMed
-
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3' end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–78. doi: 10.1016/j.molcel.2009.08.016 - DOI - PMC - PubMed
-
- Dominski Z, Carpousis AJ, Clouet-d'Orval B. Emergence of the beta-CASP ribonucleases: highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim Biophys Acta. 2013;1829:532–51. doi: 10.1016/j.bbagrm.2013.01.010 - DOI - PubMed
-
- Azzouz TN, Gruber A, Schumperli D. U7 snRNP-specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3' end processing. Nucleic Acids Res. 2005;33:2106–17. doi: 10.1093/nar/gki516 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

