Diacetylenic lipids in the design of stable lipopolymers able to complex and protect plasmid DNA
- PMID: 29020107
- PMCID: PMC5636127
- DOI: 10.1371/journal.pone.0186194
Diacetylenic lipids in the design of stable lipopolymers able to complex and protect plasmid DNA
Abstract
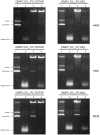
Different viral and non-viral vectors have been designed to allow the delivery of nucleic acids in gene therapy. In general, non-viral vectors have been associated with increased safety for in vivo use; however, issues regarding their efficacy, toxicity and stability continue to drive further research. Thus, the aim of this study was to evaluate the potential use of the polymerizable diacetylenic lipid 1,2-bis(10,12-tricosadiynoyl)-sn-glycero-3-phosphocholine (DC8,9PC) as a strategy to formulate stable cationic lipopolymers in the delivery and protection of plasmid DNA. Cationic lipopolymers were prepared following two different methodologies by using DC8,9PC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), and the cationic lipids (CL) 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), stearylamine (SA), and myristoylcholine chloride (MCL), in a molar ratio of 1:1:0.2 (DMPC:DC8,9PC:CL). The copolymerization methodology allowed obtaining cationic lipopolymers which were smaller in size than those obtained by the cationic addition methodology although both techniques presented high size stability over a 166-day incubation period at 4°C. Cationic lipopolymers containing DOTAP or MCL were more efficient in complexing DNA than those containing SA. Moreover, lipopolymers containing DOTAP were found to form highly stable complexes with DNA, able to resist serum DNAses degradation. Furthermore, neither of the cationic lipopolymers (with or without DNA) induced red blood cell hemolysis, although metabolic activity determined on the L-929 and Vero cell lines was found to be dependent on the cell line, the formulation and the presence of DNA. The high stability and DNA protection capacity as well as the reduced toxicity determined for the cationic lipopolymer containing DOTAP highlight the potential advantage of using lipopolymers when designing novel non-viral carrier systems for use in in vivo gene therapy. Thus, this work represents the first steps toward developing a cationic lipopolymer-based gene delivery system using polymerizable and cationic lipids.
Conflict of interest statement
Figures


References
-
- He C-X, Tabata Y, Gao J-Q. Non-viral gene delivery carrier and its three-dimensional transfection system. International journal of pharmaceutics. 2010;386(1):232–42. - PubMed
-
- Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit J-P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29(24):3477–96. - PubMed
-
- Naldini L. Gene therapy returns to centre stage. Nature. 2015;526(7573):351–60. doi: 10.1038/nature15818 - DOI - PubMed
-
- Oliveira A, Ferraz M, Monteiro F, Simoes S. Cationic liposome—DNA complexes as gene delivery vectors: development and behaviour towards bone-like cells. Acta Biomaterialia. 2009;5(6):2142–51. doi: 10.1016/j.actbio.2009.02.019 - DOI - PubMed
-
- Porras G, Bezard E. Preclinical development of gene therapy for Parkinson's disease. Experimental neurology. 2008;209(1):72–81. doi: 10.1016/j.expneurol.2007.08.003 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

