Comprehensive benchmarking of SNV callers for highly admixed tumor data
- PMID: 29020110
- PMCID: PMC5636151
- DOI: 10.1371/journal.pone.0186175
Comprehensive benchmarking of SNV callers for highly admixed tumor data
Abstract
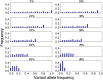
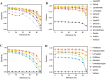
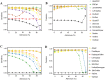
Precision medicine attempts to individualize cancer therapy by matching tumor-specific genetic changes with effective targeted therapies. A crucial first step in this process is the reliable identification of cancer-relevant variants, which is considerably complicated by the impurity and heterogeneity of clinical tumor samples. We compared the impact of admixture of non-cancerous cells and low somatic allele frequencies on the sensitivity and precision of 19 state-of-the-art SNV callers. We studied both whole exome and targeted gene panel data and up to 13 distinct parameter configurations for each tool. We found vast differences among callers. Based on our comprehensive analyses we recommend joint tumor-normal calling with MuTect, EBCall or Strelka for whole exome somatic variant calling, and HaplotypeCaller or FreeBayes for whole exome germline calling. For targeted gene panel data on a single tumor sample, LoFreqStar performed best. We further found that tumor impurity and admixture had a negative impact on precision, and in particular, sensitivity in whole exome experiments. At admixture levels of 60% to 90% sometimes seen in pathological biopsies, sensitivity dropped significantly, even when variants were originally present in the tumor at 100% allele frequency. Sensitivity to low-frequency SNVs improved with targeted panel data, but whole exome data allowed more efficient identification of germline variants. Effective somatic variant calling requires high-quality pathological samples with minimal admixture, a consciously selected sequencing strategy, and the appropriate variant calling tool with settings optimized for the chosen type of data.
Conflict of interest statement
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

