Deciphering the olfactory repertoire of the tiger mosquito Aedes albopictus
- PMID: 29020917
- PMCID: PMC5637092
- DOI: 10.1186/s12864-017-4144-1
Deciphering the olfactory repertoire of the tiger mosquito Aedes albopictus
Abstract
Background: The Asian tiger mosquito Aedes albopictus is a highly invasive species and competent vector of several arboviruses (e.g. dengue, chikungunya, Zika) and parasites (e.g. dirofilaria) of public health importance. Compared to other mosquito species, Ae. albopictus females exhibit a generalist host seeking as well as a very aggressive biting behaviour that are responsible for its high degree of nuisance. Several complex mosquito behaviours such as host seeking, feeding, mating or oviposition rely on olfactory stimuli that target a range of sensory neurons localized mainly on specialized head appendages such as antennae, maxillary palps and the mouthparts.
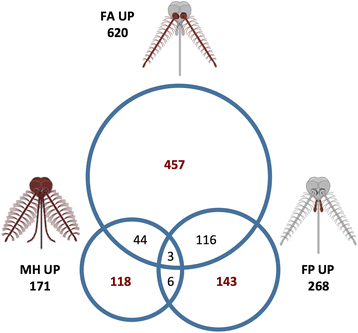
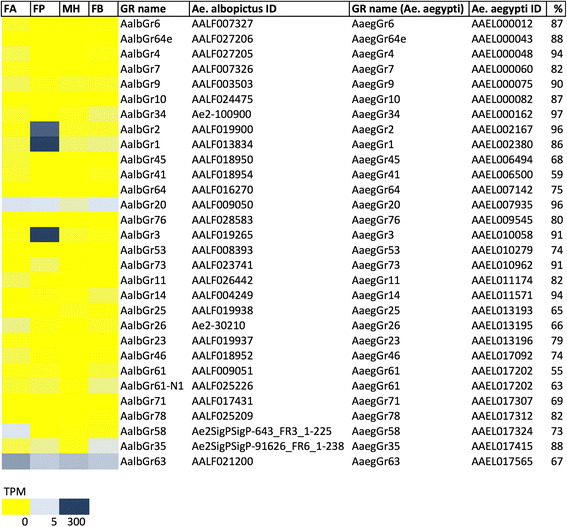
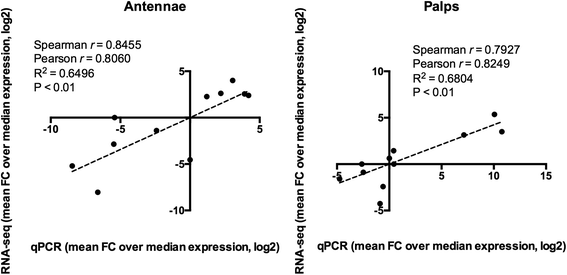
Results: With the aim to describe the Ae. albopictus olfactory repertoire we have used RNA-seq to reveal the transcriptome profiles of female antennae and maxillary palps. Male heads and whole female bodies were employed as reference for differential expression analysis. The relative transcript abundance within each tissue (TPM, transcripts per kilobase per million) and the pairwise differential abundance in the different tissues (fold change values and false discovery rates) were evaluated. Contigs upregulated in the antennae (620) and maxillary palps (268) were identified and relative GO and PFAM enrichment profiles analysed. Chemosensory genes were described: overall, 77 odorant binding proteins (OBP), 82 odorant receptors (OR), 60 ionotropic receptors (IR) and 30 gustatory receptors (GR) were identified by comparative genomics and transcriptomics. In addition, orthologs of genes expressed in the female/male maxillary palps and/or antennae and involved in thermosensation (e.g. pyrexia and arrestin1), mechanosensation (e.g. piezo and painless) and neuromodulation were classified.
Conclusions: We provide here the first detailed transcriptome of the main Ae. albopictus sensory appendages, i.e. antennae and maxillary palps. A deeper knowledge of the olfactory repertoire of the tiger mosquito will help to better understand its biology and may pave the way to design new attractants/repellents.
Keywords: Aedes albopictus; Antennae; Chemosensory genes; Maxillary palps; Mosquito; Olfaction; mRNA-sequencing.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

