SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis
- PMID: 29020972
- PMCID: PMC5637259
- DOI: 10.1186/s12943-017-0724-6
SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis
Abstract
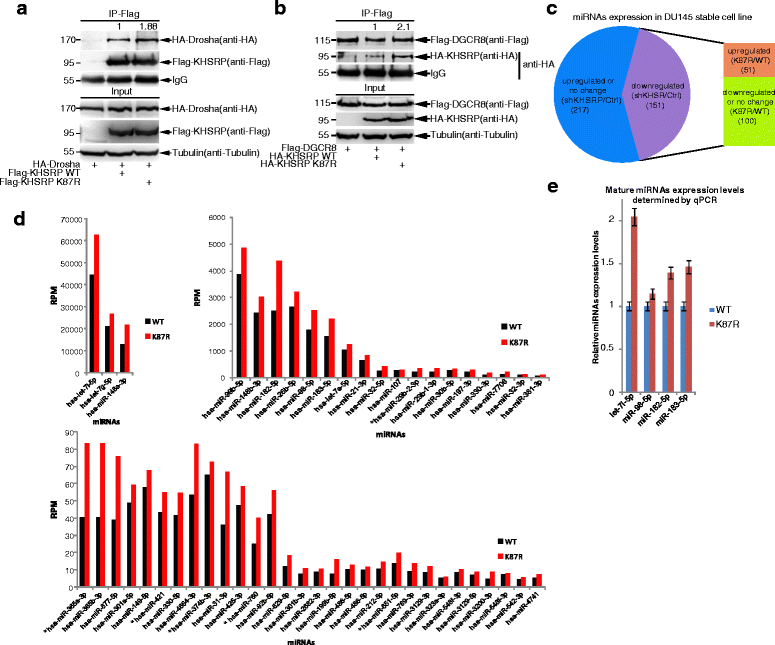
Background: MicroRNAs (miRNAs) are important regulators involved in diverse physiological and pathological processes including cancer. SUMO (small ubiquitin-like modifier) is a reversible protein modifier. We recently found that SUMOylation of TARBP2 and DGCR8 is involved in the regulation of the miRNA pathway. KHSRP is a single stranded nucleic acid binding protein with roles in transcription and mRNA decay, and it is also a component of the Drosha-DGCR8 complex promoting the miRNA biogenesis.
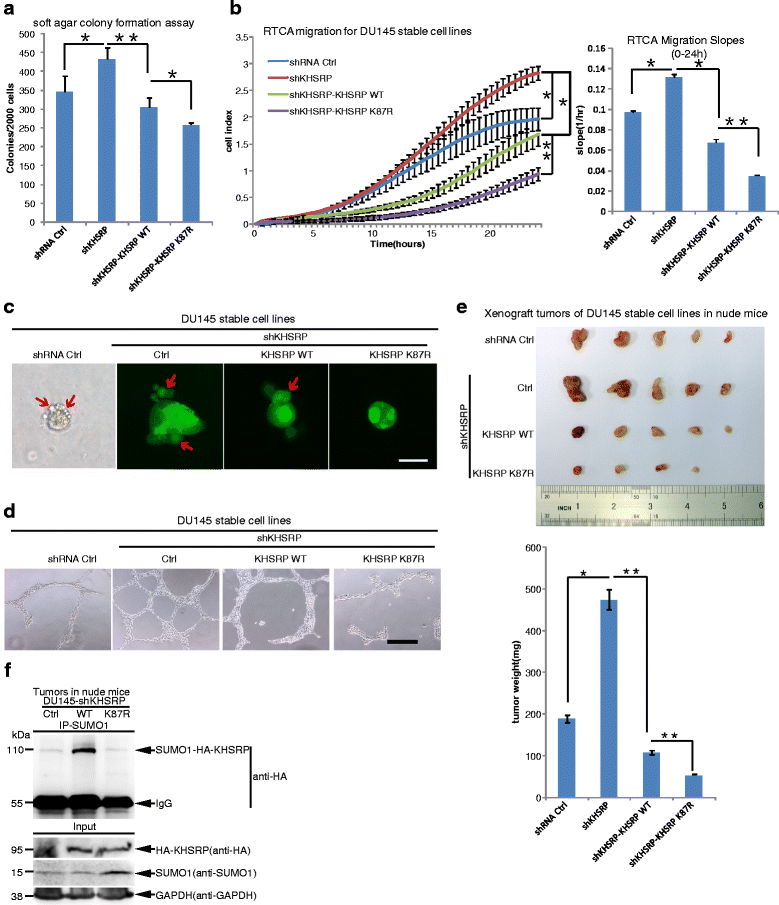
Methods: The in vivo SUMOylation assay using the Ni2+-NTA affinity pulldown or immunoprecipitation (IP) and the in vitro E.coli-based SUMOylation assay were used to analyze SUMOylation of KHSRP. Nuclear/Cytosol fractionation assay and immunofluorescent staining were used to observe the localization of KHSRP. High-throughput miRNA sequencing, quantantive RT-PCR and RNA immunoprecipitation assay (RIP) were employed to determine the effects of KHSRP SUMO1 modification on the miRNA biogenesis. The soft-agar colony formation, migration, vasculogenic mimicry (VM) and three-dimensional (3D) cell culture assays were performed to detect the phenotypes of tumor cells in vitro, and the xenograft tumor model in mice was conducted to verify that SUMO1 modification of KHSRP regulated tumorigenesis in vivo.
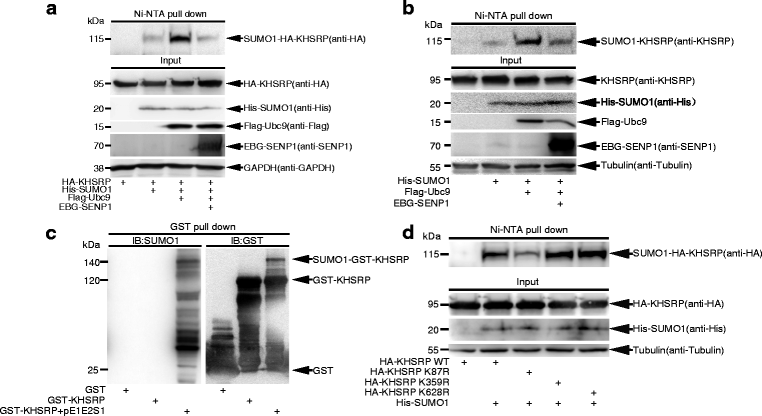
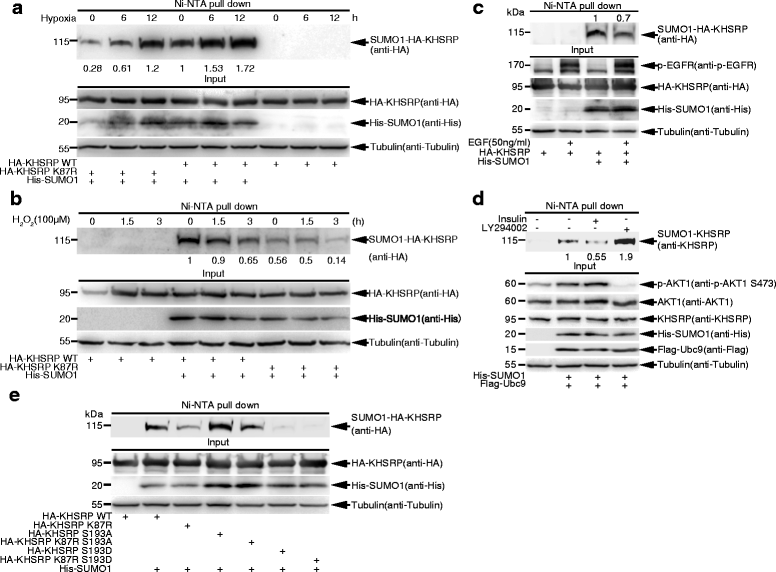
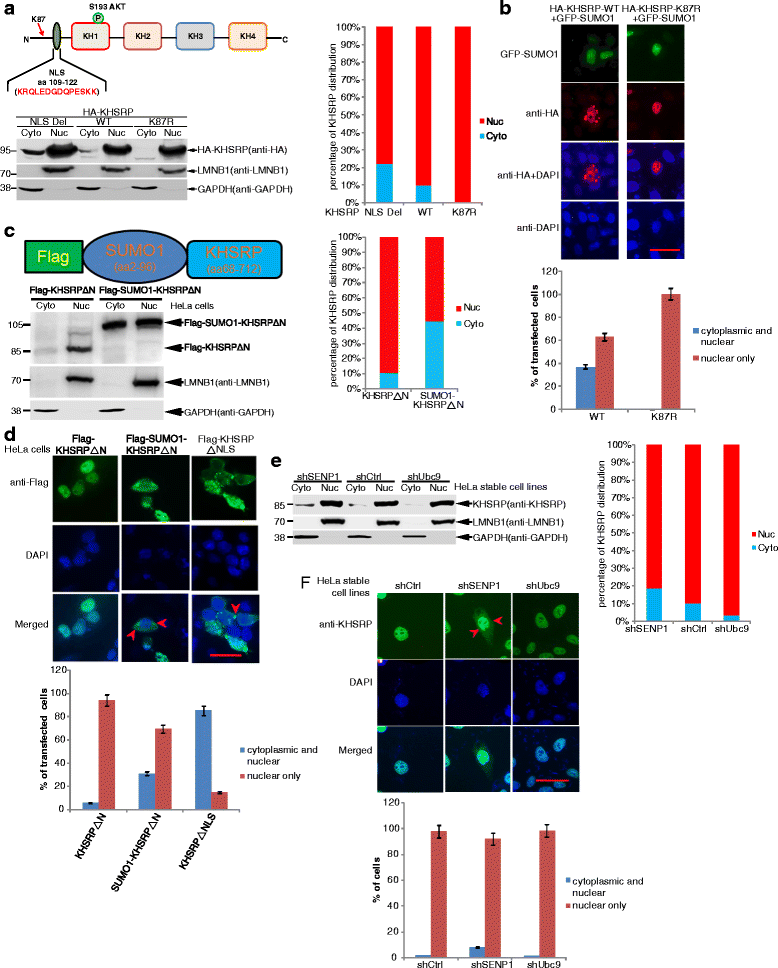
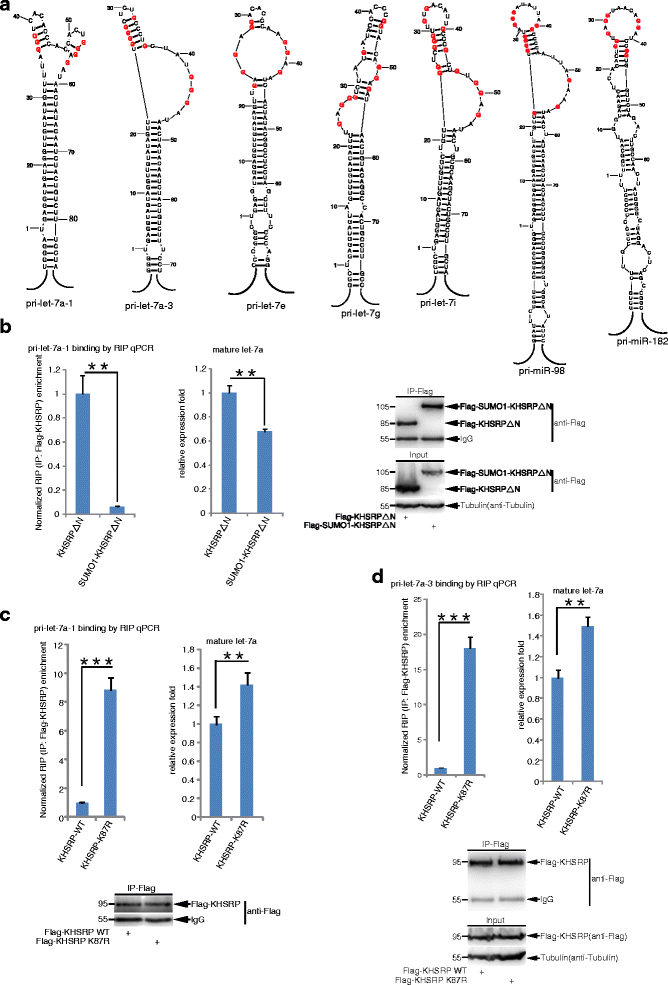
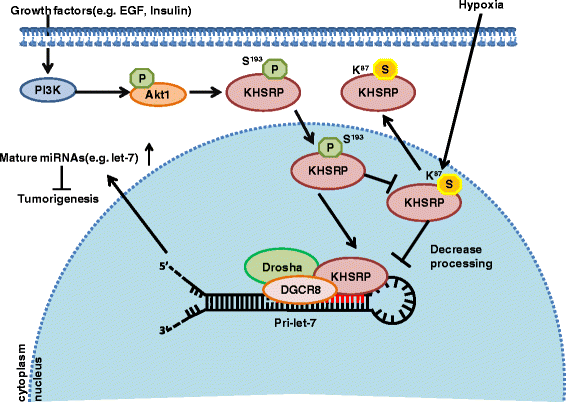
Results: KHSRP is modified by SUMO1 at the major site K87, and this modification can be increased upon the microenvironmental hypoxia while reduced by the treatment with growth factors. SUMO1 modification of KHSRP inhibits its interaction with the pri-miRNA/Drosha-DGCR8 complex and probably increases its translocation from the nucleus to the cytoplasm. Consequently, SUMO1 modification of KHSRP impairs the processing step of pre-miRNAs from pri-miRNAs which especially harbor short G-rich stretches in their terminal loops (TL), resulting in the downregulation of a subset of TL-G-Rich miRNAs such as let-7 family and consequential tumorigenesis.
Conclusions: Our data demonstrate how the miRNA biogenesis pathway is connected to tumorigenesis and cancer progression through the reversible SUMO1 modification of KHSRP.
Keywords: KHSRP; SUMO1 modification; TL-G-Rich miRNA biogenesis; Tumorgenesis.
Conflict of interest statement
Ethics approval
All animal studies were conducted with the approval and guidance of Shanghai Jiao Tong University Medical Animal Ethics Committees.
Consent for publication
All authors read and approved the final manuscript, and consent to publish.
Competing interests
The authors declare that they have no competing interests.
Figures
Similar articles
-
SUMOylation at K707 of DGCR8 controls direct function of primary microRNA.Nucleic Acids Res. 2015 Sep 18;43(16):7945-60. doi: 10.1093/nar/gkv741. Epub 2015 Jul 21. Nucleic Acids Res. 2015. PMID: 26202964 Free PMC article.
-
The Ubiquitin-specific Protease USP36 Associates with the Microprocessor Complex and Regulates miRNA Biogenesis by SUMOylating DGCR8.Cancer Res Commun. 2023 Mar 20;3(3):459-470. doi: 10.1158/2767-9764.CRC-22-0344. eCollection 2023 Mar. Cancer Res Commun. 2023. PMID: 36950067 Free PMC article.
-
SUMOylation of Grb2 enhances the ERK activity by increasing its binding with Sos1.Mol Cancer. 2014 Apr 29;13:95. doi: 10.1186/1476-4598-13-95. Mol Cancer. 2014. PMID: 24775912 Free PMC article.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
-
Terminal loop-mediated control of microRNA biogenesis.Biochem Soc Trans. 2012 Aug;40(4):789-93. doi: 10.1042/BST20120053. Biochem Soc Trans. 2012. PMID: 22817735 Review.
Cited by
-
SUMOylation of PUM2 promotes the vasculogenic mimicry of glioma cells via regulating CEBPD.Clin Transl Med. 2020 Sep;10(5):e168. doi: 10.1002/ctm2.168. Clin Transl Med. 2020. PMID: 32997416 Free PMC article.
-
KHSRP has oncogenic functions and regulates the expression and alternative splicing of DNA repair genes in breast cancer MDA-MB-231 cells.Sci Rep. 2024 Jun 26;14(1):14694. doi: 10.1038/s41598-024-64687-0. Sci Rep. 2024. PMID: 38926398 Free PMC article.
-
Membrane-cytoplasm translocation of annexin A4 is involved in the metastasis of colorectal carcinoma.Aging (Albany NY). 2021 Mar 24;13(7):10312-10325. doi: 10.18632/aging.202793. Epub 2021 Mar 24. Aging (Albany NY). 2021. PMID: 33761465 Free PMC article.
-
The RNA-binding protein KSRP aggravates malignant progression of clear cell renal cell carcinoma through transcriptional inhibition and post-transcriptional destabilization of the NEDD4L ubiquitin ligase.J Biomed Sci. 2023 Aug 14;30(1):68. doi: 10.1186/s12929-023-00949-9. J Biomed Sci. 2023. PMID: 37580757 Free PMC article.
-
Distinctive tumorigenic significance and innovative oncology targets of SUMOylation.Theranostics. 2024 May 19;14(8):3127-3149. doi: 10.7150/thno.97162. eCollection 2024. Theranostics. 2024. PMID: 38855173 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

