Histone Acetyltransferase KAT6A Upregulates PI3K/AKT Signaling through TRIM24 Binding
- PMID: 29021135
- PMCID: PMC5690809
- DOI: 10.1158/0008-5472.CAN-17-1388
Histone Acetyltransferase KAT6A Upregulates PI3K/AKT Signaling through TRIM24 Binding
Abstract
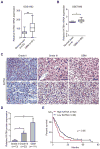
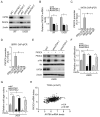
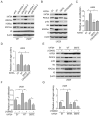
Lysine acetyltransferase KAT6A is a chromatin regulator that contributes to histone modification and cancer, but the basis of its actions are not well understood. Here, we identify a KAT6A signaling pathway that facilitates glioblastoma (GBM), where it is upregulated. KAT6A expression was associated with GBM patient survival. KAT6A silencing suppressed cell proliferation, cell migration, colony formation, and tumor development in an orthotopic mouse xenograft model system. Mechanistic investigations demonstrated that KAT6A acetylates lysine 23 of histone H3 (H3K23), which recruits the nuclear receptor binding protein TRIM24 to activate PIK3CA transcription, thereby enhancing PI3K/AKT signaling and tumorigenesis. Overexpressing activated AKT or PIK3CA rescued the growth inhibition due to KAT6A silencing. Conversely, the pan-PI3K inhibitor LY294002 abrogated the growth-promoting effect of KAT6A. Overexpression of KAT6A or TRIM24, but not KAT6A acetyltransferase activity-deficient mutants or TRIM24 mutants lacking H3K23ac-binding sites, promoted PIK3CA expression, AKT phosphorylation, and cell proliferation. Taken together, our results define an essential role of KAT6A in glioma formation, rationalizing its candidacy as a therapeutic target for GBM treatment. Cancer Res; 77(22); 6190-201. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. - PubMed
-
- Maleszewska M, Kaminska B. Deregulation of histone-modifying enzymes and chromatin structure modifiers contributes to glioma development. Future oncology. 2015;11:2587–601. - PubMed
-
- Waldmann T, Schneider R. Targeting histone modifications--epigenetics in cancer. Current opinion in cell biology. 2013;25:184–9. - PubMed
-
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. - PubMed
-
- Voss AK, Collin C, Dixon MP, Thomas T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Developmental cell. 2009;17:674–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

