A microtubule polymerase cooperates with the kinesin-6 motor and a microtubule cross-linker to promote bipolar spindle assembly in the absence of kinesin-5 and kinesin-14 in fission yeast
- PMID: 29021344
- PMCID: PMC5706992
- DOI: 10.1091/mbc.E17-08-0497
A microtubule polymerase cooperates with the kinesin-6 motor and a microtubule cross-linker to promote bipolar spindle assembly in the absence of kinesin-5 and kinesin-14 in fission yeast
Abstract
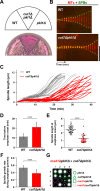
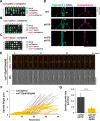
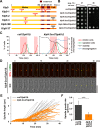
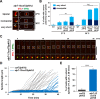
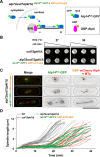
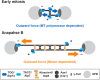
Accurate chromosome segregation relies on the bipolar mitotic spindle. In many eukaryotes, spindle formation is driven by the plus-end-directed motor kinesin-5 that generates outward force to establish spindle bipolarity. Its inhibition leads to the emergence of monopolar spindles with mitotic arrest. Intriguingly, simultaneous inactivation of the minus-end-directed motor kinesin-14 restores spindle bipolarity in many systems. Here we show that in fission yeast, three independent pathways contribute to spindle bipolarity in the absence of kinesin-5/Cut7 and kinesin-14/Pkl1. One is kinesin-6/Klp9 that engages with spindle elongation once short bipolar spindles assemble. Klp9 also ensures the medial positioning of anaphase spindles to prevent unequal chromosome segregation. Another is the Alp7/TACC-Alp14/TOG microtubule polymerase complex. Temperature-sensitive alp7cut7pkl1 mutants are arrested with either monopolar or very short spindles. Forced targeting of Alp14 to the spindle pole body is sufficient to render alp7cut7pkl1 triply deleted cells viable and promote spindle assembly, indicating that Alp14-mediated microtubule polymerization from the nuclear face of the spindle pole body could generate outward force in place of Cut7 during early mitosis. The third pathway involves the Ase1/PRC1 microtubule cross-linker that stabilizes antiparallel microtubules. Our study, therefore, unveils multifaceted interplay among kinesin-dependent and -independent pathways leading to mitotic bipolar spindle assembly.
© 2017 Yukawa et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

Similar articles
-
Kinesin-5-independent mitotic spindle assembly requires the antiparallel microtubule crosslinker Ase1 in fission yeast.Nat Commun. 2017 May 17;8:15286. doi: 10.1038/ncomms15286. Nat Commun. 2017. PMID: 28513584 Free PMC article.
-
Kinesin-6 Klp9 plays motor-dependent and -independent roles in collaboration with Kinesin-5 Cut7 and the microtubule crosslinker Ase1 in fission yeast.Sci Rep. 2019 May 14;9(1):7336. doi: 10.1038/s41598-019-43774-7. Sci Rep. 2019. PMID: 31089172 Free PMC article.
-
Two spatially distinct kinesin-14 proteins, Pkl1 and Klp2, generate collaborative inward forces against kinesin-5 Cut7 in S. pombe.J Cell Sci. 2018 Jan 4;131(1):jcs210740. doi: 10.1242/jcs.210740. J Cell Sci. 2018. PMID: 29167352
-
[Kinesin-14 leaps to pole position in bipolar spindle assembly].Ai Zheng. 2008 Sep;27(9):989-92. Ai Zheng. 2008. PMID: 18799042 Review. Chinese.
-
How Essential Kinesin-5 Becomes Non-Essential in Fission Yeast: Force Balance and Microtubule Dynamics Matter.Cells. 2020 May 7;9(5):1154. doi: 10.3390/cells9051154. Cells. 2020. PMID: 32392819 Free PMC article. Review.
Cited by
-
The Putative RNA-Binding Protein Dri1 Promotes the Loading of Kinesin-14/Klp2 to the Mitotic Spindle and Is Sequestered into Heat-Induced Protein Aggregates in Fission Yeast.Int J Mol Sci. 2021 Apr 30;22(9):4795. doi: 10.3390/ijms22094795. Int J Mol Sci. 2021. PMID: 33946513 Free PMC article.
-
Antiparallel microtubule bundling supports KIF15-driven mitotic spindle assembly.Mol Biol Cell. 2024 Jun 1;35(6):ar84. doi: 10.1091/mbc.E24-01-0023. Epub 2024 Apr 10. Mol Biol Cell. 2024. PMID: 38598297 Free PMC article.
-
CLASP promotes microtubule bundling in metaphase spindle independently of Ase1/PRC1 in fission yeast.Biol Open. 2019 Oct 24;8(10):bio045716. doi: 10.1242/bio.045716. Biol Open. 2019. PMID: 31615768 Free PMC article.
-
Kinesin-6 Klp9 orchestrates spindle elongation by regulating microtubule sliding and growth.Elife. 2021 Jun 3;10:e67489. doi: 10.7554/eLife.67489. Elife. 2021. PMID: 34080538 Free PMC article.
-
Fission yeast Ase1PRC1 is required for the G2-microtubule damage response.Mol Biol Res Commun. 2021 Dec;10(4):179-188. doi: 10.22099/mbrc.2021.41001.1650. Mol Biol Res Commun. 2021. PMID: 35097140 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

