The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella
- PMID: 29021545
- PMCID: PMC5636795
- DOI: 10.1038/s41467-017-00889-7
The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella
Abstract
The enzyme fructose-bisphosphate aldolase occupies a central position in glycolysis and gluconeogenesis pathways. Beyond its housekeeping role in metabolism, fructose-bisphosphate aldolase has been involved in additional functions and is considered as a potential target for drug development against pathogenic bacteria. Here, we address the role of fructose-bisphosphate aldolase in the bacterial pathogen Francisella novicida. We demonstrate that fructose-bisphosphate aldolase is important for bacterial multiplication in macrophages in the presence of gluconeogenic substrates. In addition, we unravel a direct role of this metabolic enzyme in transcription regulation of genes katG and rpoA, encoding catalase and an RNA polymerase subunit, respectively. We propose a model in which fructose-bisphosphate aldolase participates in the control of host redox homeostasis and the inflammatory immune response.The enzyme fructose-bisphosphate aldolase (FBA) plays central roles in glycolysis and gluconeogenesis. Here, Ziveri et al. show that FBA of the pathogen Francisella novicida acts, in addition, as a transcriptional regulator and is important for bacterial multiplication in macrophages.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
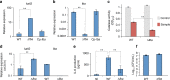


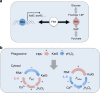
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

