A cdk1 gradient guides surface contraction waves in oocytes
- PMID: 29021609
- PMCID: PMC5636809
- DOI: 10.1038/s41467-017-00979-6
A cdk1 gradient guides surface contraction waves in oocytes
Erratum in
-
Publisher Correction: A cdk1 gradient guides surface contraction waves in oocytes.Nat Commun. 2018 Jan 10;9(1):200. doi: 10.1038/s41467-017-02520-1. Nat Commun. 2018. PMID: 29321627 Free PMC article.
Abstract
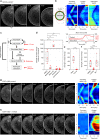
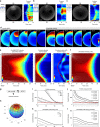
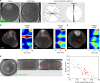
Surface contraction waves (SCWs) in oocytes and embryos lead to large-scale shape changes coupled to cell cycle transitions and are spatially coordinated with the cell axis. Here, we show that SCWs in the starfish oocyte are generated by a traveling band of myosin II-driven cortical contractility. At the front of the band, contractility is activated by removal of cdk1 inhibition of the RhoA/RhoA kinase/myosin II signaling module, while at the rear, contractility is switched off by negative feedback originating downstream of RhoA kinase. The SCW's directionality and speed are controlled by a spatiotemporal gradient of cdk1-cyclinB. This gradient is formed by the release of cdk1-cyclinB from the asymmetrically located nucleus, and progressive degradation of cyclinB. By combining quantitative imaging, biochemical and mechanical perturbations with mathematical modeling, we demonstrate that the SCWs result from the spatiotemporal integration of two conserved regulatory modules, cdk1-cyclinB for cell cycle regulation and RhoA/Rok/NMYII for actomyosin contractility.Surface contraction waves (SCWs) are prominent shape changes coupled to cell cycle transitions in oocytes. Here the authors show that SCWs are patterned by the spatiotemporal integration of two conserved modules, cdk1-cyclinB for cell cycle regulation and RhoA/Rok/NMYII for actomyosin contractility.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Yoneda M, Kobayakawa Y, Kubota HY, Sakai M. Surface contraction waves in amphibian eggs. J. Cell. Sci. 1982;54:35–46. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

