Intracellular Crosslinking of Filoviral Nucleoproteins with Xintrabodies Restricts Viral Packaging
- PMID: 29021793
- PMCID: PMC5623874
- DOI: 10.3389/fimmu.2017.01197
Intracellular Crosslinking of Filoviral Nucleoproteins with Xintrabodies Restricts Viral Packaging
Abstract
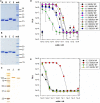
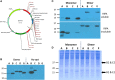
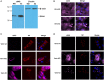
Viruses assemble large macromolecular repeat structures that become part of the infectious particles or virions. Ribonucleocapsids (RNCs) of negative strand RNA viruses are a prime example where repetition of nucleoprotein (NP) along the genome creates a core polymeric helical scaffold that accommodates other nucleocapsid proteins including viral polymerase. The RNCs are transported through the cytosol for packaging into virions through association with viral matrix proteins at cell membranes. We hypothesized that RNC would be ideal targets for crosslinkers engineered to promote aberrant protein-protein interactions, thereby blocking their orderly transport and packaging. Previously, we had generated single-domain antibodies (sdAbs) against Filoviruses that have all targeted highly conserved C-terminal regions of NP known to be repetitively exposed along the length of the RNCs of Marburgvirus (MARV) and Ebolavirus (EBOV). Our crosslinker design consisted of dimeric sdAb expressed intracellularly, which we call Xintrabodies (X- for crosslinking). Electron microscopy of purified NP polymers incubated with purified sdAb constructs showed NP aggregation occurred in a genus-specific manner with dimeric and not monomeric sdAb. A virus-like particle (VLP) assay was used for initial evaluation where we found that dimeric sdAb inhibited NP incorporation into VP40-based VLPs whereas monomeric sdAb did not. Inhibition of NP packaging was genus specific. Confocal microscopy revealed dimeric sdAb was diffuse when expressed alone but focused on pools of NP when the two were coexpressed, while monomeric sdAb showed ambivalent partition. Infection of stable Vero cell lines expressing dimeric sdAb specific for either MARV or EBOV NP resulted in smaller plaques and reduced progeny of cognate virus relative to wild-type Vero cells. Though the impact was marginal at later time-points, the collective data suggest that viral replication can be reduced by crosslinking intracellular NP using relatively small amounts of dimeric sdAb to restrict NP packaging. The stoichiometry and ease of application of the approach would likely benefit from transitioning away from intracellular expression of crosslinking sdAb to exogenous delivery of antibody. By retuning sdAb specificity, the approach of crosslinking highly conserved regions of assembly critical proteins may well be applicable to inhibiting replication processes of a broad spectrum of viruses.
Keywords: Ebola; Marburg; VHH; crosslinker; intrabody; nucleoprotein; single-domain antibodies; virus-like particle.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

