A new and improved algorithm for the quantification of chromatin condensation from microscopic data shows decreased chromatin condensation in regenerating axolotl limb cells
- PMID: 29023511
- PMCID: PMC5638231
- DOI: 10.1371/journal.pone.0185292
A new and improved algorithm for the quantification of chromatin condensation from microscopic data shows decreased chromatin condensation in regenerating axolotl limb cells
Abstract
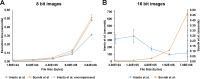
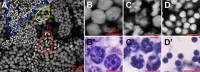
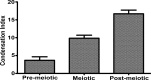
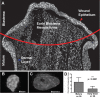
The nuclear landscape plays an important role in the regulation of tissue and positional specific genes in embryonic and developing cells. Changes in this landscape can be dynamic, and are associated with the differentiation of cells during embryogenesis, and the de-differentiation of cells during induced pluripotent stem cell (iPSC) formation and in many cancers. However, tools to quantitatively characterize these changes are limited, especially in the in vivo context, where numerous tissue types are present and cells are arranged in multiple layers. Previous tools have been optimized for the monolayer nature of cultured cells. Therefore, we present a new algorithm to quantify the condensation of chromatin in two in vivo systems. We first developed this algorithm to quantify changes in chromatin compaction and validated it in differentiating spermatids in zebrafish testes. Our algorithm successfully detected the typical increase in chromatin compaction as these cells differentiate. We then employed the algorithm to quantify the changes that occur in amphibian limb cells as they participate in a regenerative response. We observed that the chromatin in the limb cells de-compacts as they contribute to the regenerating organ. We present this new tool as an open sourced software that can be readily accessed and optimized to quantify chromatin compaction in complex multi-layered samples.
Conflict of interest statement
Figures
References
-
- Chromatin Structure: A Repeating Unit of Histones and DNA. American Association for the Advancement of Science; 1974;184: 868–871. doi: 10.1126/science.184.4139.868 - DOI - PubMed
-
- Crystal structure of the nucleosome core particle at 2.8|[thinsp]||[Aring]| resolution. Nature Publishing Group; 1997;389: 251–260. doi: 10.1038/38444 - DOI - PubMed
-
- Translating the Histone Code. American Association for the Advancement of Science; 2001;293: 1074–1080. doi: 10.1126/science.1063127 - DOI - PubMed
-
- Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. American Association for the Advancement of Science; 2002;298: 1039–1043. doi: 10.1126/science.1076997 - DOI - PubMed
-
- Ezh1 and Ezh2 Maintain Repressive Chromatin through Different Mechanisms. Elsevier Inc; 2008;32: 503–518. doi: 10.1016/j.molcel.2008.11.004 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

