Enhanced Stem Cell Differentiation and Immunopurification of Genome Engineered Human Retinal Ganglion Cells
- PMID: 29024560
- PMCID: PMC6430043
- DOI: 10.1002/sctm.17-0059
Enhanced Stem Cell Differentiation and Immunopurification of Genome Engineered Human Retinal Ganglion Cells
Abstract
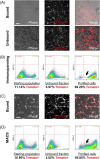
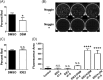
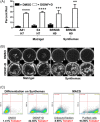
Human pluripotent stem cells have the potential to promote biological studies and accelerate drug discovery efforts by making possible direct experimentation on a variety of human cell types of interest. However, stem cell cultures are generally heterogeneous and efficient differentiation and purification protocols are often lacking. Here, we describe the generation of clustered regularly-interspaced short palindromic repeats(CRISPR)-Cas9 engineered reporter knock-in embryonic stem cell lines in which tdTomato and a unique cell-surface protein, THY1.2, are expressed under the control of the retinal ganglion cell (RGC)-enriched gene BRN3B. Using these reporter cell lines, we greatly improved adherent stem cell differentiation to the RGC lineage by optimizing a novel combination of small molecules and established an anti-THY1.2-based protocol that allows for large-scale RGC immunopurification. RNA-sequencing confirmed the similarity of the stem cell-derived RGCs to their endogenous human counterparts. Additionally, we developed an in vitro axonal injury model suitable for studying signaling pathways and mechanisms of human RGC cell death and for high-throughput screening for neuroprotective compounds. Using this system in combination with RNAi-based knockdown, we show that knockdown of dual leucine kinase (DLK) promotes survival of human RGCs, expanding to the human system prior reports that DLK inhibition is neuroprotective for murine RGCs. These improvements will facilitate the development and use of large-scale experimental paradigms that require numbers of pure RGCs that were not previously obtainable. Stem Cells Translational Medicine 2017;6:1972-1986.
Keywords: Biotechnology; Cell differentiation; Clustered regularly interspaced short palindromic repeats; Retinal ganglion cells; Stem cells.
© 2017 The Authors Stem Cells Translational Medicine published by Wiley Periodicals, Inc. on behalf of AlphaMed Press.
Conflict of interest statement
The authors indicated no potential conflicts of interest.
Figures




References
-
- Levin LA, Gordon LK. Retinal ganglion cell disorders: Types and treatments. Prog Retin Eye Res 2002;21:465–484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

