Mevalonate 5-diphosphate mediates ATP binding to the mevalonate diphosphate decarboxylase from the bacterial pathogen Enterococcus faecalis
- PMID: 29025876
- PMCID: PMC5766736
- DOI: 10.1074/jbc.M117.802223
Mevalonate 5-diphosphate mediates ATP binding to the mevalonate diphosphate decarboxylase from the bacterial pathogen Enterococcus faecalis
Abstract
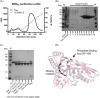
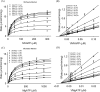
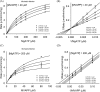
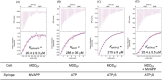
The mevalonate pathway produces isopentenyl diphosphate (IPP), a building block for polyisoprenoid synthesis, and is a crucial pathway for growth of the human bacterial pathogen Enterococcus faecalis The final enzyme in this pathway, mevalonate diphosphate decarboxylase (MDD), acts on mevalonate diphosphate (MVAPP) to produce IPP while consuming ATP. This essential enzyme has been suggested as a therapeutic target for the treatment of drug-resistant bacterial infections. Here, we report functional and structural studies on the mevalonate diphosphate decarboxylase from E. faecalis (MDDEF). The MDDEF crystal structure in complex with ATP (MDDEF-ATP) revealed that the phosphate-binding loop (amino acids 97-105) is not involved in ATP binding and that the phosphate tail of ATP in this structure is in an outward-facing position pointing away from the active site. This suggested that binding of MDDEF to MVAPP is necessary to guide ATP into a catalytically favorable position. Enzymology experiments show that the MDDEF performs a sequential ordered bi-substrate reaction with MVAPP as the first substrate, consistent with the isothermal titration calorimetry (ITC) experiments. On the basis of ITC results, we propose that this initial prerequisite binding of MVAPP enhances ATP binding. In summary, our findings reveal a substrate-induced substrate-binding event that occurs during the MDDEF-catalyzed reaction. The disengagement of the phosphate-binding loop concomitant with the alternative ATP-binding configuration may provide the structural basis for antimicrobial design against these pathogenic enterococci.
Keywords: Enterococcus; crystal structure; decarboxylase; drug resistance; enzyme kinetics; enzyme mechanism; isothermal titration calorimetry (ITC); mevalonate pathway.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- World Health Organization (2014) Antimicrobial Resistance: Global Report on Surveillance 2014
-
- Centers for Disease Control and Prevention (2013) Antibiotic Resistance Threats in the United States, Report 2013
-
- Paulsen I. T., Banerjei L., Myers G. S., Nelson K. E., Seshadri R., Read T. D., Fouts D. E., Eisen J. A., Gill S. R., Heidelberg J. F., Tettelin H., Dodson R. J., Umayam L., Brinkac L., Beanan M., et al. (2003) Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299, 2071–2074 - PubMed
-
- Uttley A. H., Collins C. H., Naidoo J., and George R. C. (1988) Vancomycin-resistant enterococci. Lancet 1, 57–58 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

