Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass
- PMID: 29025995
- PMCID: PMC5748897
- DOI: 10.1126/science.aam9305
Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass
Abstract
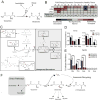
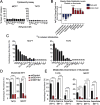
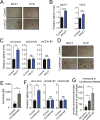
Ammonia is a ubiquitous by-product of cellular metabolism; however, the biological consequences of ammonia production are not fully understood, especially in cancer. We found that ammonia is not merely a toxic waste product but is recycled into central amino acid metabolism to maximize nitrogen utilization. In our experiments, human breast cancer cells primarily assimilated ammonia through reductive amination catalyzed by glutamate dehydrogenase (GDH); secondary reactions enabled other amino acids, such as proline and aspartate, to directly acquire this nitrogen. Metabolic recycling of ammonia accelerated proliferation of breast cancer. In mice, ammonia accumulated in the tumor microenvironment and was used directly to generate amino acids through GDH activity. These data show that ammonia is not only a secreted waste product but also a fundamental nitrogen source that can support tumor biomass.
Copyright © 2017, American Association for the Advancement of Science.
Figures

Comment in
-
Feeding frenzy for cancer cells.Science. 2017 Nov 17;358(6365):862-863. doi: 10.1126/science.aaq1070. Science. 2017. PMID: 29146793 No abstract available.
-
When Cancer Cells Are Given Lemo[NH3]s, They Make Lemo[NH3]ade.Cell Metab. 2017 Dec 5;26(6):811-813. doi: 10.1016/j.cmet.2017.11.008. Cell Metab. 2017. PMID: 29211980
References
-
- Coloff JL, et al. Differential Glutamate Metabolism in Proliferating and Quiescent Mammary Epithelial Cells. Cell metabolism. 2016;23:867–880. - PubMed
-
- Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Science signaling. 2010;3:ra31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

