Crystal structure of the flexible tandem repeat domain of bacterial cellulose synthesis subunit C
- PMID: 29026093
- PMCID: PMC5638805
- DOI: 10.1038/s41598-017-12530-0
Crystal structure of the flexible tandem repeat domain of bacterial cellulose synthesis subunit C
Abstract
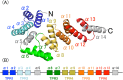
Bacterial cellulose (BC) is synthesized and exported through the cell membrane via a large protein complex (terminal complex) that consists of three or four subunits. BcsC is a little-studied subunit considered to export BC to the extracellular matrix. It is predicted to have two domains: a tetratrico peptide repeat (TPR) domain and a β-barrelled outer membrane domain. Here we report the crystal structure of the N-terminal part of BcsC-TPR domain (Asp24-Arg272) derived from Enterobacter CJF-002. Unlike most TPR-containing proteins which have continuous TPR motifs, this structure has an extra α-helix between two clusters of TPR motifs. Five independent molecules in the crystal had three different conformations that varied at the hinge of the inserted α-helix. Such structural feature indicates that the inserted α-helix confers flexibility to the chain and changes the direction of the TPR super-helix, which was also suggested by structural analysis of BcsC-TPR (Asp24-Leu664) in solution by size exclusion chromatography-small-angle X-ray scattering. The flexibility at the α-helical hinge may play important role for exporting glucan chains.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

