Genetic and Epigenetic Profiling Reveals EZH2-mediated Down Regulation of OCT-4 Involves NR2F2 during Cardiac Differentiation of Human Embryonic Stem Cells
- PMID: 29026152
- PMCID: PMC5638931
- DOI: 10.1038/s41598-017-13442-9
Genetic and Epigenetic Profiling Reveals EZH2-mediated Down Regulation of OCT-4 Involves NR2F2 during Cardiac Differentiation of Human Embryonic Stem Cells
Abstract
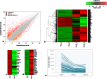
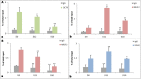
Human embryonic (hES) stem cells are widely used as an in vitro model to understand global genetic and epigenetic changes that occur during early embryonic development. In-house derived hES cells (KIND1) were subjected to directed differentiation into cardiovascular progenitors (D12) and beating cardiomyocytes (D20). Transcriptome profiling of undifferentiated (D0) and differentiated (D12 and 20) cells was undertaken by microarray analysis. ChIP and sequential ChIP were employed to study role of transcription factor NR2F2 during hES cells differentiation. Microarray profiling showed that an alteration of about 1400 and 1900 transcripts occurred on D12 and D20 respectively compared to D0 whereas only 19 genes were altered between D12 and D20. This was found associated with corresponding expression pattern of chromatin remodelers, histone modifiers, miRNAs and lncRNAs marking the formation of progenitors and cardiomyocytes on D12 and D20 respectively. ChIP sequencing and sequential ChIP revealed the binding of NR2F2 with polycomb group member EZH2 and pluripotent factor OCT4 indicating its crucial involvement in cardiac differentiation. The study provides a detailed insight into genetic and epigenetic changes associated with hES cells differentiation into cardiac cells and a role for NR2F2 is deciphered for the first time to down-regulate OCT-4 via EZH2 during cardiac differentiation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

