Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches
- PMID: 29026658
- PMCID: PMC5610714
- DOI: 10.1099/mgen.0.000118
Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches
Erratum in
-
Corrigendum: Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches.Microb Genom. 2018 Mar;4(3):e000130. doi: 10.1099/mgen.0.000130. Microb Genom. 2018. PMID: 31192781 Free PMC article. No abstract available.
Abstract
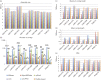
This study aimed to assess the feasibility of using the Oxford Nanopore Technologies (ONT) MinION long-read sequencer in reconstructing fully closed plasmid sequences from eight Enterobacteriaceae isolates of six different species with plasmid populations of varying complexity. Species represented were Escherichia coli, Klebsiella pneumoniae, Citrobacter freundii, Enterobacter cloacae, Serratia marcescens and Klebsiella oxytoca, with plasmid populations ranging from 1-11 plasmids with sizes of 2-330 kb. Isolates were sequenced using Illumina (short-read) and ONT's MinION (long-read) platforms, and compared with fully resolved PacBio (long-read) sequence assemblies for the same isolates. We compared the performance of different assembly approaches including SPAdes, plasmidSPAdes, hybridSPAdes, Canu, Canu+Pilon (canuPilon) and npScarf in recovering the plasmid structures of these isolates by comparing with the gold-standard PacBio reference sequences. Overall, canuPilon provided consistently good quality assemblies both in terms of assembly statistics (N50, number of contigs) and assembly accuracy [presence of single nucleotide polymorphisms (SNPs)/indels with respect to the reference sequence]. For plasmid reconstruction, Canu recovered 70 % of the plasmids in complete contigs, and combining three assembly approaches (Canu or canuPilon, hybridSPAdes and plasmidSPAdes) resulted in a total 78 % recovery rate for all the plasmids. The analysis demonstrated the potential of using MinION sequencing technology to resolve important plasmid structures in Enterobacteriaceae species independent of and in conjunction with Illumina sequencing data. A consensus assembly derived from several assembly approaches could present significant benefit in accurately resolving the greatest number of plasmid structures.
Keywords: Gram-negative Enterobactericacae; MinION nanopore sequencing; plasmid assembly; plasmid reconstruction.
Figures


References
-
- Mathers AJ, Stoesser N, Sheppard AE, Pankhurst L, Giess A, et al. Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae at a single institution: insights into endemicity from whole-genome sequencing. Antimicrob Agents Chemother. 2015;59:1656–1663. doi: 10.1128/AAC.04292-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

