Functional characterization of novel NR5A1 variants reveals multiple complex roles in disorders of sex development
- PMID: 29027299
- PMCID: PMC5765430
- DOI: 10.1002/humu.23354
Functional characterization of novel NR5A1 variants reveals multiple complex roles in disorders of sex development
Abstract
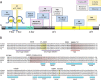
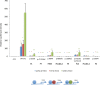
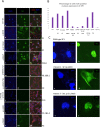
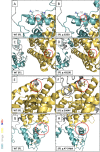
Variants in the NR5A1 gene encoding SF1 have been described in a diverse spectrum of disorders of sex development (DSD). Recently, we reported the use of a targeted gene panel for DSD where we identified 15 individuals with a variant in NR5A1, nine of which are novel. Here, we examine the functional effect of these changes in relation to the patient phenotype. All novel variants tested had reduced trans-activational activity, while several had altered protein level, localization, or conformation. In addition, we found evidence of new roles for SF1 protein domains including a region within the ligand binding domain that appears to contribute to SF1 regulation of Müllerian development. There was little correlation between the severity of the phenotype and the nature of the NR5A1 variant. We report two familial cases of NR5A1 deficiency with evidence of variable expressivity; we also report on individuals with oligogenic inheritance. Finally, we found that the nature of the NR5A1 variant does not inform patient outcomes (including pubertal androgenization and malignancy risk). This study adds nine novel pathogenic NR5A1 variants to the pool of diagnostic variants. It highlights a greater need for understanding the complexity of SF1 function and the additional factors that contribute.
Keywords: NR5A1; disorders of sex development; genotype-phenotype correlation; mutation; oligogenic; variable expressivity.
© 2017 The Authors. Human Mutation published by Wiley Periodicals, Inc.
Figures

References
-
- Achermann, J. C. , Ito, M. , Ito, M. , Hindmarsh, P. , & Jameson, J. L. 1999. A mutation in the gene encoding steroidogenic factor‐1 causes XY sex reversal and adrenal failure in humans. Nature Genetics, 22, 125–126. - PubMed
-
- Allali, S. , Muller, J‐B. , Brauner, R. , Lourenço, D. , Boudjenah, R. , Karageorgou, V. , … Bashamboo, A. (2011). Mutation analysis of NR5A1 encoding steroidogenic factor 1 in 77 patients with 46, XY disorders of sex development (DSD) including hypospadias. PLoS One, 6, e24117–e2418. - PMC - PubMed
-
- Arango, N. A. , Lovell‐Badge, R. , & Behringer, R. R. (1999). Targeted mutagenesis of the endogenous mouse. Cell, 99, 409–419. - PubMed
-
- Baetens, D. , Mladenov, W. , Delle Chiaie, B. , Menten, B. , Desloovere, A. , Iotova, V. , … Cools, M. (2014). Extensive clinical, hormonal and genetic screening in a large consecutive series of 46,XY neonates and infants with atypical sexual development. Orphanet Journal of Rare Diseases, 9, 209. - PMC - PubMed
-
- Barbaro, M. , Cools, M. , Looijenga, L. H. J. , Drop, S. L. S. , & Wedell, A. (2011). Partial deletion of the NR5A1 (SF1) gene detected by synthetic probe MLPA in a patient with XY gonadal disorder of sex development. Sexual Development, 5, 181–187. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

