Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions
- PMID: 29028171
- PMCID: PMC5682607
- DOI: 10.1021/acs.jmedchem.7b01221
Structure-Based Design of Non-natural Macrocyclic Peptides That Inhibit Protein-Protein Interactions
Abstract
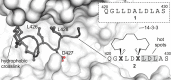
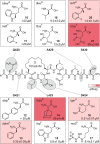
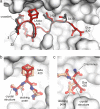
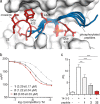
Macrocyclic peptides can interfere with challenging biomolecular targets including protein-protein interactions. Whereas there are various approaches that facilitate the identification of peptide-derived ligands, their evolution into higher affinity binders remains a major hurdle. We report a virtual screen based on molecular docking that allows the affinity maturation of macrocyclic peptides taking non-natural amino acids into consideration. These macrocycles bear large and flexible substituents that usually complicate the use of docking approaches. A virtual library containing more than 1400 structures was screened against the target focusing on docking poses with the core structure resembling a known bioactive conformation. Based on this screen, a macrocyclic peptide 22 involving two non-natural amino acids was evolved showing increased target affinity and biological activity. Predicted binding modes were verified by X-ray crystallography. The presented workflow allows the screening of large macrocyclic peptides with diverse modifications thereby expanding the accessible chemical space and reducing synthetic efforts.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

