An Analysis of the Epidemic of Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae: Convergence of Two Evolutionary Mechanisms Creates the "Perfect Storm"
- PMID: 29029188
- PMCID: PMC5853647
- DOI: 10.1093/infdis/jix524
An Analysis of the Epidemic of Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae: Convergence of Two Evolutionary Mechanisms Creates the "Perfect Storm"
Abstract
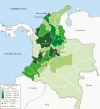
Background: Carbapenem resistance is a critical healthcare challenge worldwide. Particularly concerning is the widespread dissemination of Klebsiella pneumoniae carbapenemase (KPC). Klebsiella pneumoniae harboring blaKPC (KPC-Kpn) is endemic in many areas including the United States, where the epidemic was primarily mediated by the clonal dissemination of Kpn ST258. We postulated that the spread of blaKPC in other regions occurs by different and more complex mechanisms. To test this, we investigated the evolution and dynamics of spread of KPC-Kpn in Colombia, where KPC became rapidly endemic after emerging in 2005.
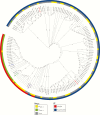
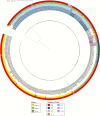
Methods: We sequenced the genomes of 133 clinical isolates recovered from 24 tertiary care hospitals located in 10 cities throughout Colombia, between 2002 (before the emergence of KPC-Kpn) and 2014. Phylogenetic reconstructions and evolutionary mapping were performed to determine temporal and genetic associations between the isolates.
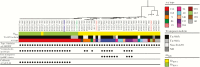
Results: Our results indicate that the start of the epidemic was driven by horizontal dissemination of mobile genetic elements carrying blaKPC-2, followed by the introduction and subsequent spread of clonal group 258 (CG258) isolates containing blaKPC-3.
Conclusions: The combination of 2 evolutionary mechanisms of KPC-Kpn within a challenged health system of a developing country created the "perfect storm" for sustained endemicity of these multidrug-resistant organisms in Colombia.
Keywords: Colombia; KPC; Klebsiella pneumoniae.
© The Author 2017. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures

References
-
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics.Geneva, Switzerland: World Health Organization; 2017.
-
- Diene SM, Rolain JM. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin Microbiol Infect 2014; 20:831–8. - PubMed
-
- Logan LK. Carbapenem-resistant enterobacteriaceae: an emerging problem in children. Clin Infect Dis 2012; 55:852–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

