GATA2 regulates the erythropoietin receptor in t(12;21) ALL
- PMID: 29029492
- PMCID: PMC5630392
- DOI: 10.18632/oncotarget.19792
GATA2 regulates the erythropoietin receptor in t(12;21) ALL
Abstract
The t(12;21) (p13;q22) chromosomal translocation resulting in the ETV6/RUNX1 fusion gene is the most frequent structural cytogenetic abnormality in children with acute lymphoblastic leukemia (ALL). The erythropoietin receptor (EPOR), usually associated with erythroid progenitor cells, is highly expressed in ETV6/RUNX1 positive cases compared to other B-lineage ALL subtypes. Gene expression analysis of a microarray database and direct quantitative analysis of patient samples revealed strong correlation between EPOR and GATA2 expression in ALL, and higher expression of GATA2 in t(12;21) patients. The mechanism of EPOR regulation was mainly investigated using two B-ALL cell lines: REH, which harbor and express the ETV6/RUNX1 fusion gene; and NALM-6, which do not. Expression of EPOR was increased in REH cells compared to NALM-6 cells. Moreover, of the six GATA family members only GATA2 was differentially expressed with substantially higher levels present in REH cells. GATA2 was shown to bind to the EPOR 5'-UTR in REH, but did not bind in NALM-6 cells. Overexpression of GATA2 led to an increase in EPOR expression in REH cells only, indicating that GATA2 regulates EPOR but is dependent on the cellular context. Both EPOR and GATA2 are hypomethylated and associated with increased mRNA expression in REH compared to NALM-6 cells. Decitabine treatment effectively reduced methylation of CpG sites in the GATA2 promoter leading to increased GATA2 expression in both cell lines. Although Decitabine also reduced an already low level of methylation of the EPOR in NALM-6 cells there was no increase in EPOR expression. Furthermore, EPOR and GATA2 are regulated post-transcriptionally by miR-362 and miR-650, respectively. Overall our data show that EPOR expression in t(12;21) B-ALL cells, is regulated by GATA2 and is mediated through epigenetic, transcriptional and post-transcriptional mechanisms, contingent upon the genetic subtype of the disease.
Keywords: EPOR; GATA2; acute lymphoblastic leukemia; epigenetic; transcriptional regulation.
Figures
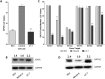



 ), the EPOR transcription start site (TCSS), the EPOR translation start site (TLSS) and the CpG sites included in the pyrosequencing assay (grey dashed box). All genomic coordinates are given relative to the TCSS. (B) Schematic of the GATA2 genomic locus showing the relative positions of CpG dinucleotides (
), the EPOR transcription start site (TCSS), the EPOR translation start site (TLSS) and the CpG sites included in the pyrosequencing assay (grey dashed box). All genomic coordinates are given relative to the TCSS. (B) Schematic of the GATA2 genomic locus showing the relative positions of CpG dinucleotides ( ), the GATA2 transcription start site (TCSS) and the CpG sites included in the pyrosequencing assay (grey dashed box). (C)
EPOR 5′ DNA specific pyrosequencing assays were performed on bisulphite converted DNA prepared from REH, NALM-6 and UT-7 cells. DNA methylation was assessed at 18 CpG sites in triplicate. Whiskers indicate Tukey minimum and maximum CpG methylation values; boxes indicate inter-quartile range, with the median marked. Significant enrichments were detected by one-way ANOVA and are indicated by *** (p < 0.001). (D)
GATA2 5′ DNA specific pyrosequencing assays were performed on bisulphite converted DNA prepared from REH, NALM-6 and UT-7 cells. DNA methylation was assessed at 5 CpG sites in triplicate. Whiskers indicate Tukey minimum and maximum CpG methylation values; boxes indicate inter-quartile range, with the median marked. Significant enrichments were detected by one-way ANOVA and are indicated by *** (p < 0.001).
), the GATA2 transcription start site (TCSS) and the CpG sites included in the pyrosequencing assay (grey dashed box). (C)
EPOR 5′ DNA specific pyrosequencing assays were performed on bisulphite converted DNA prepared from REH, NALM-6 and UT-7 cells. DNA methylation was assessed at 18 CpG sites in triplicate. Whiskers indicate Tukey minimum and maximum CpG methylation values; boxes indicate inter-quartile range, with the median marked. Significant enrichments were detected by one-way ANOVA and are indicated by *** (p < 0.001). (D)
GATA2 5′ DNA specific pyrosequencing assays were performed on bisulphite converted DNA prepared from REH, NALM-6 and UT-7 cells. DNA methylation was assessed at 5 CpG sites in triplicate. Whiskers indicate Tukey minimum and maximum CpG methylation values; boxes indicate inter-quartile range, with the median marked. Significant enrichments were detected by one-way ANOVA and are indicated by *** (p < 0.001).
References
-
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–43. - PubMed
-
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, Liu HC, Mahfouz R, Raimondi SC, Lenny N, Patel A, Downing JR. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–9. https://doi.org/10.1182/blood-2003-01-0338. - DOI - PubMed
-
- Fine BM, Stanulla M, Schrappe M, Ho M, Viehmann S, Harbott J, Boxer LM. Gene expression patterns associated with recurrent chromosomal translocations in acute lymphoblastic leukemia. Blood. 2004;103:1043–9. https://doi.org/10.1182/blood-2003-05-1518. - DOI - PubMed
-
- Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, Horstmann MA, Janka-Schaub GE, Kamps WA, et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–34. https://doi.org/10.1016/S1470-2045(08)70339-5. - DOI - PMC - PubMed
-
- Inthal A, Krapf G, Beck D, Joas R, Kauer MO, Orel L, Fuka G, Mann G, Panzer-Grumayer ER. Role of the erythropoietin receptor in ETV6/RUNX1-positive acute lymphoblastic leukemia. Clin Cancer Res. 2008;14:7196–204. https://doi.org/10.1158/1078-0432.CCR-07-5051. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

