Structural insights into marine carbohydrate degradation by family GH16 κ-carrageenases
- PMID: 29030427
- PMCID: PMC5712629
- DOI: 10.1074/jbc.M117.808279
Structural insights into marine carbohydrate degradation by family GH16 κ-carrageenases
Abstract
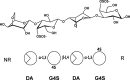
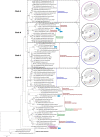
Carrageenans are sulfated α-1,3-β-1,4-galactans found in the cell wall of some red algae that are practically valuable for their gelation and biomimetic properties but also serve as a potential carbon source for marine bacteria. Carbohydrate degradation has been studied extensively for terrestrial plant/bacterial systems, but sulfation is not present in these cases, meaning the marine enzymes used to degrade carrageenans must possess unique features to recognize these modifications. To gain insights into these features, we have focused on κ-carrageenases from two distant bacterial phyla, which belong to glycoside hydrolase family 16 and cleave the β-1,4 linkage of κ-carrageenan. We have solved the crystal structure of the catalytic module of ZgCgkA from Zobellia galactanivorans at 1.66 Å resolution and compared it with the only other structure available, that of PcCgkA from Pseudoalteromonas carrageenovora 9T (ATCC 43555T). We also describe the first substrate complex in the inactivated mutant form of PcCgkA at 1.7 Å resolution. The structural and biochemical comparison of these enzymes suggests key determinants that underlie the functional properties of this subfamily. In particular, we identified several arginine residues that interact with the polyanionic substrate, and confirmed the functional relevance of these amino acids using a targeted mutagenesis strategy. These results give new insight into the diversity of the κ-carrageenase subfamily. The phylogenetic analyses show the presence of several distinct clades of enzymes that relate to differences in modes of action or subtle differences within the same substrate specificity, matching the hybrid character of the κ-carrageenan polymer.
Keywords: carbohydrate recognition; crystal structure; enzyme kinetics; enzyme-substrate complex; kappa-carrageenan; polysaccharide; processivity; site-directed mutagenesis; sulfated polysaccharides.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- O'Dowd C. D., Jimenez J. L., Bahreini R., Flagan R. C., Seinfeld J. H., Hämeri K., Pirjola L., Kulmala M., Jennings S. G., and Hoffmann T. (2002) Marine aerosol formation from biogenic iodine emissions. Nature 417, 632–636 - PubMed
-
- Duggins D. O., Simenstad C. A., and Estes J. A. (1989) Magnification of secondary production by kelp detritus in coastal marine ecosystems. Science 245, 170–173 - PubMed
-
- Martin M., Portetelle D., Michel G., and Vandenbol M. (2014) Microorganisms living on macroalgae: diversity, interactions, and biotechnological applications. Appl. Microbiol. Biotechnol. 98, 2917–2935 - PubMed
-
- Larsbrink J., Rogers T. E., Hemsworth G. R., McKee L. S., Tauzin A. S., Spadiut O., Klinter S., Pudlo N. A., Urs K., Koropatkin N. M., Creagh A. L., Haynes C. A., Kelly A. G., Cederholm S. N., Davies G. J., Martens E. C., and Brumer H. (2014) A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

