Direct binding to integrins and loss of disulfide linkage in interleukin-1β (IL-1β) are involved in the agonistic action of IL-1β
- PMID: 29030430
- PMCID: PMC5723996
- DOI: 10.1074/jbc.M117.818302
Direct binding to integrins and loss of disulfide linkage in interleukin-1β (IL-1β) are involved in the agonistic action of IL-1β
Abstract
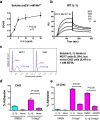
There is a strong link between integrins and interleukin-1β (IL-1β), but the specifics of the role of integrins in IL-1β signaling are unclear. We describe that IL-1β specifically bound to integrins αvβ3 and α5β1. The E128K mutation in the IL1R-binding site enhanced integrin binding. We studied whether direct integrin binding is involved in IL-1β signaling. We compared sequences of IL-1β and IL-1 receptor antagonist (IL1RN), which is an IL-1β homologue but has no agonistic activity. Several surface-exposed Lys residues are present in IL-1β, but not in IL1RN. A disulfide linkage is present in IL1RN, but is not in IL-1β because of natural C117F mutation. Substitution of the Lys residues to Glu markedly reduced integrin binding of E128K IL-1β, suggesting that the Lys residues mediate integrin binding. The Lys mutations reduced, but did not completely abrogate, agonistic action of IL-1β. We studied whether the disulfide linkage plays a role in agonistic action of IL-1β. Reintroduction of the disulfide linkage by the F117C mutation did not affect agonistic activity of WT IL-1β, but effectively reduced the remaining agonistic activity of the Lys mutants. Also, deletion of the disulfide linkage in IL1RN by the C116F mutation did not make it agonistic. We propose that the direct binding to IL-1β to integrins is primarily important for agonistic IL-1β signaling, and that the disulfide linkage indirectly affects signaling by blocking conformational changes induced by weak integrin binding to the Lys mutants. The integrin-IL-1β interaction is a potential target for drug discovery.
Keywords: IL-1β; NF-κB (NF-KB); agonistic action; cell signaling; disulfide linkage; integrin; interleukin-1 (IL-1); mutagenesis.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

