RBPJ/CBF1 interacts with L3MBTL3/MBT1 to promote repression of Notch signaling via histone demethylase KDM1A/LSD1
- PMID: 29030483
- PMCID: PMC5666606
- DOI: 10.15252/embj.201796525
RBPJ/CBF1 interacts with L3MBTL3/MBT1 to promote repression of Notch signaling via histone demethylase KDM1A/LSD1
Abstract
Notch signaling is an evolutionarily conserved signal transduction pathway that is essential for metazoan development. Upon ligand binding, the Notch intracellular domain (NOTCH ICD) translocates into the nucleus and forms a complex with the transcription factor RBPJ (also known as CBF1 or CSL) to activate expression of Notch target genes. In the absence of a Notch signal, RBPJ acts as a transcriptional repressor. Using a proteomic approach, we identified L3MBTL3 (also known as MBT1) as a novel RBPJ interactor. L3MBTL3 competes with NOTCH ICD for binding to RBPJ In the absence of NOTCH ICD, RBPJ recruits L3MBTL3 and the histone demethylase KDM1A (also known as LSD1) to the enhancers of Notch target genes, leading to H3K4me2 demethylation and to transcriptional repression. Importantly, in vivo analyses of the homologs of RBPJ and L3MBTL3 in Drosophila melanogaster and Caenorhabditis elegans demonstrate that the functional link between RBPJ and L3MBTL3 is evolutionarily conserved, thus identifying L3MBTL3 as a universal modulator of Notch signaling in metazoans.
Keywords: RBPJ; KDM1A; L3MBTL3; Notch signaling.
© 2017 The Authors. Published under the terms of the CC BY NC ND 4.0 license.
Figures

Detection of the RBPJ/L3MBTL3 interaction using the yeast two‐hybrid (Y2H) assay. In this Y2H experiment, RBPJ is fused to the GAL4 DNA‐binding (DB) domain and L3MBTL3 is fused to the GAL4 activation domain (AD). The DB‐RBPJ and AD‐L3MBTL3 fusion proteins interact with each other, leading to the activation of the ADE2 and HIS3 reporter genes and allowing yeast cells to grow on selective media lacking adenine or histidine. The six Y2H controls were previously described (Dreze et al, 2010). The experiment was independently replicated thrice.
Endogenous L3MBTL3 co‐purifies specifically with HA‐RBPJ but not with HA‐EGFP, HA‐TBL1X, or HA‐HEY2. Immuno‐precipitation (IP) of HA‐tagged RBPJ, EGFP, TBL1X, or HEY2 in U87‐MG cells followed by Western blot analyses using HA or L3MBTL3 antibody. The experiment was independently replicated twice.
Endogenous RBPJ co‐purifies specifically with HA‐L3MBTL3 but not with HA‐EGFP, HA‐TBL1X, or HA‐HEY2. IPs of HA‐tagged L3MBTL3, EGFP, TBL1X, or HEY2 in U87‐MG cells followed by Western blot analyses using HA or RBPJ antibody. The experiment was independently replicated twice.

Schematic representation of the L3MBTL3 protein and the deletion mutants used in panel (B). The L3MBTL3 protein (XP_006715641.1) consists of a C2C2 zinc finger (ZnF #1; CDD: 128717), three MBT domains (CDD: 214723), a C2H2 zinc finger (ZnF #2; CDD: 201844), and a sterile α motif domain (SAM; CDD: 197735).
L3MBTL3‐Δ(1‐64) does not interact with RBPJ. IP of HA‐FLAG‐tagged RBPJ in the presence of FLAG‐tagged L3MBTL3 (WT or deletion mutants) in HEK293T cells followed by Western blotting using FLAG antibody. The experiment was independently replicated twice.
Schematic representation of the RBPJ protein and the deletion mutants used in panels (D and E). The RBPJ protein (XP_005248218.1) consists of the N‐terminal domain (NTD), the β‐trefoil domain (BTD), and the C‐terminal domain (CTD).
Deletion of the BTD domain impairs the RBPJ/L3MBTL3 interaction. IP of HA‐tagged L3MBTL3 in the presence of FLAG‐tagged RBPJ (WT and deletion mutants) in HEK293T cells followed by Western blotting using HA or FLAG antibody. The experiment was independently replicated twice.
RBPJF261R point mutant does not interact with L3MBTL3. IP of HA‐tagged L3MBTL3 in the presence of FLAG‐tagged RBPJ (WT and point mutants) in HEK293T cells followed by Western blotting using HA or FLAG antibody. RBPJV263R and RBPJA284R also show a reduced ability to interact with L3MBTL3. The experiment was independently replicated twice.

- A
Thermodynamic characterization of the RBPJ/L3MBTL3 interaction. Representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for L3MBTL3‐(31‐70) binding to RBPJ‐(53‐474).
- B, C
NOTCH1 ICD outcompetes L3MBTL3 for binding to RBPJ in a dose‐dependent manner. IPs were performed in CRISPR/Cas9‐mediated L3MBTL3 knockout (KO) HEK293T cells. (B) SBP‐FLAG‐RBPJ and HA‐L3MBTL3‐Δ(SAM) in the presence of an increasing amount of HA‐NOTCH1 ICD. (C) SBP‐FLAG‐RBPJ and HA‐NOTCH1 ICD in the presence of an increasing amount of HA‐L3MBTL3‐Δ(SAM). The L3MBTL3‐Δ(SAM) mutant construct was used instead of the L3MBTL3 WT construct in order to allow the analysis of both NOTCH1 ICD and L3MBTL3 proteins in the same Western blot. CRISPR/Cas9 sg‐L3MBTL3‐resistant plasmids were used to express HA‐L3MBTL3‐Δ(SAM). The experiment was independently replicated thrice. WB, Western blot; IP, immuno‐precipitation.
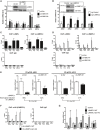
De‐repression of Notch target genes upon RBPJ knockdown. Shown are means ± s.d. of quadruplicate experiments. *P < 0.05, **P < 0.01, NS, not significant; one‐way ANOVA model on log‐transformed data. Inset: Western blot analysis validates the shRNA‐mediated depletion of RBPJ.
De‐repression of Notch target genes in L3MBTL3 KO U87‐MG cells. Shown are means ± s.d. of quadruplicate experiments. **P < 0.01, NS, not significant; two‐sample t‐test on log‐transformed data. Inset: Western blot analysis validates the CRISPR/Cas9‐mediated KO of L3MBTL3.
RBPJ and L3MBTL3 co‐localize at the proximal Notch‐responsive elements of Notch target genes. Shown are means ± s.d. of triplicate ChIP experiments.
L3MBTL3 occupancy at the proximal Notch‐responsive elements of Notch target genes decreases upon RBPJ knockdown. Shown are means ± s.d. of triplicate ChIP experiments.
The repressive activity of L3MBTL3 at Notch target genes is RBPJ dependent. Expression analysis of Notch target genes upon RBPJ knockdown and/or overexpression of L3MBTL3. Shown are means ± s.d. of triplicate experiments. P‐values were estimated via a one‐way ANOVA model on log‐transformed data where the difference of differences was tested, which is equivalent to testing the interaction in a two‐way ANOVA model. Western blot analysis validates the overexpression of L3MBTL3 and the shRNA‐mediated depletion of RBPJ (Appendix Fig S3E). Gene expression analyses of OCT4 was performed as control (Appendix Fig S3F).
L3MBTL3 occupancy at the proximal Notch‐responsive elements of Notch target genes is dependent on its RBPJ‐interacting domain. ChIP analyses of HA‐L3MBTL3 WT and HA‐L3MBTL3‐Δ(1‐64) occupancy at the proximal Notch‐responsive elements of Notch target genes. Shown are means ± s.d. of duplicate experiments measured twice each.
The L3MBTL3‐(1‐64) domain is required for the downregulation of HES1 and HEY2 in U87‐MG cells. Expression analysis of Notch target genes upon overexpression of L3MBTL3 WT, L3MBTL3‐Δ(1‐64), or LacZ control (Control). Shown are means ± s.d. of triplicate experiments. *P < 0.05, **P < 0.01, NS, not significant; one‐way ANOVA model on log‐transformed data.
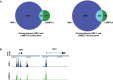
Venn diagram showing the genomewide co‐localization of RBPJ and L3MBTL3.
Snapshots showing the co‐localization of RBPJ and L3MBTL3 at the HES1 and HEY2 loci.

L3MBTL3 and KDM1A interact in yeast two‐hybrid assay. In this Y2H experiment, KDM1A is fused to the GAL4 DNA‐binding (DB) domain and L3MBTL3 is fused to the GAL4 activation domain (AD). The DB‐KDM1A and AD‐L3MBTL3 fusion proteins interact with each other, leading to the activation of the HIS3 reporter gene and allowing yeast cells to grow on selective media lacking histidine. The six Y2H controls have been previously described (Dreze et al, 2010). The experiment was independently replicated thrice.
Endogenous RBPJ interacts with both endogenous KDM1A and endogenous L3MBTL3. IP of RBPJ in U87‐MG or MDA‐MB‐231 cells using a RBPJ antibody followed by Western blot analyses using KDM1A, L3MBTL3, or RBPJ antibody. The experiment was independently replicated twice.
L3MBTL3 interacts with KDM1A in IP experiments. L3MBTL3 KO U87‐MG cells were transfected with CRISPR/Cas9 sg‐L3MBTL3‐resistant plasmids encoding V5‐L3MBTL3 WT or V5‐L3MBTL3‐Δ(1‐64) mutant. IPs were performed using V5 antibody and the precipitates were analyzed via Western blotting using V5, KDM1A, or RBPJ antibody. The experiment was independently replicated twice.
Endogenous KDM1A interacts with FLAG‐HA‐tagged RBPJ. IP of FLAG‐HA‐tagged RBPJ in U87‐MG cells followed by Western blot analyses using RBPJ or KDM1A antibody. The experiment was independently replicated twice.
Endogenous RBPJ interacts with FLAG‐HA‐tagged KDM1A. IP of FLAG‐HA‐tagged KDM1A in U87‐MG cells followed by Western blotting analyses using FLAG or RBPJ antibody. The experiment was independently replicated twice.
The SAM domain of L3MBTL3 is required for the L3MBTL3/KDM1A interaction. HEK293T cells were transfected with HA‐tagged KDM1A and FLAG‐tagged L3MBTL3 (WT or mutants, represented in Fig 2A). Upon IP with HA antibody, proteins were analyzed via Western blotting using HA or L3MBTL3 antibody. The experiment was independently replicated twice.

The RBPJ/KDM1A interaction is indirect and occurs via L3MBTL3. IP of HA‐KDM1A in the presence of overexpressed V5‐L3MBTL3 or V5‐L3MBTL3‐Δ(1‐64) in L3MBTL3 KO U87‐MG cells. CRISPR/Cas9 sg‐L3MBTL3‐resistant plasmids were used to overexpress the L3MBTL3 proteins. The experiment was independently replicated twice.
KDM1A occupancy at the proximal Notch‐responsive elements of Notch target genes is L3MBTL3 dependent. ChIP analysis of endogenous KDM1A in L3MBTL3 KO U87‐MG cells. Shown are means ± s.d. of duplicate experiments measured twice each.
KDM1A occupancy at the proximal Notch‐responsive elements of Notch target genes is dependent on L3MBTL3, and both its RBPJ interaction and KDM1A interaction domains. ChIP analysis of endogenous KDM1A in L3MBTL3 KO U87‐MG cells upon overexpression of L3MBTL3, L3MBTL3‐Δ(1‐64) or L3MBTL3‐Δ(SAM). Control: empty vector. Shown are means ± s.d. of duplicate experiments measured twice each.
L3MBTL3, but neither L3MBTL3‐Δ(1‐64) nor L3MBTL3‐Δ(SAM), leads to decreasing H3K4me2 at the proximal Notch‐responsive element of HES1. ChIP analysis of H3K4me2 at the proximal Notch‐responsive element of HES1 upon overexpression of LacZ control (Control), L3MBTL3, L3MBTL3‐Δ(1‐64), or L3MBTL3‐Δ(SAM) in L3MBTL3 KO U87‐MG cells. Shown are means ± s.d. of duplicate experiments measured twice each. P‐values were estimated via a one‐way ANOVA on log‐transformed data.
L3MBTL3, but neither L3MBTL3‐Δ(1‐64) nor L3MBTL3‐Δ(SAM), represses HES1. Expression analysis of HES1 upon overexpression of LacZ control (Control), L3MBTL3, L3MBTL3‐Δ(1‐64), or L3MBTL3‐Δ(SAM) mutants in L3MBTL3 KO U87‐MG cells. Shown are means ± s.d. of triplicate experiments. P‐values were estimated via a one‐way ANOVA on log‐transformed data. NS, not significant. WB, Western blot; IP, immuno‐precipitation. We note that in the context of this experiment, that is, in the absence of endogenous L3MBTL3, the overexpression of L3MBTL3‐Δ(1‐64) does not result in the increased expression of HES1, contrasting with the result obtained in Fig 4G, that is, in the presence of endogenous L3MBTL3. Indeed, as expected, the dominant negative effect of L3MBTL3‐(1‐64) on endogenous WT L3MBTL3's ability to repress the expression of Notch target genes can only be observed when WT L3MBTL3 is expressed.

GST pulldown showing that dL(3)mbt, the Drosophila homolog of L3MBTL3, directly interacts with Su(H), the Drosophila homolog of RBPJ. In vitro‐transcribed and translated dL(3)mbt or dNotch ICD (dNotch ICD fragment containing the RAM domain and ANK repeats), as positive control, was incubated with bacterially purified GST‐Su(H) or GST alone pre‐bound to GSH beads. Proteins were resolved via SDS–PAGE and signals were acquired via X‐ray exposure. The experiment was independently replicated four times.
dL(3)mbt and Su(H) co‐localize genomewide. Venn diagram showing the genomewide co‐localization of dL(3)mbt and Su(H).
Snapshot showing the co‐localization of dL(3)mbt and Su(H) at the dNotch (N) locus.
In the wing imaginal disk, dL(3)mbt overexpression in the dorso–ventral (D‐V) boundary results in the downregulation of the Notch target gene cut. Wing disks expressing UAS‐GFP (top panels) or UAS‐HA‐dL(3)mbt;UAS‐GFP (bottom panels) under the vg‐Gal4 driver at 25°C were stained for cut and HA. GFP marks the vg‐Gal4 expression domain. Insets below each panel show a closer view of the D‐V boundary with yellow arrows marking the regions where HA‐dL(3)mbt is expressed and cut is downregulated. At least 20 disks for each genotype were analyzed. Representative images are shown. Scale bars: 100 μm.
The vg‐Gal4‐driven HA‐dL(3)mbt overexpression causes a serrated wing (wing notching) phenotype. Flies expressing either UAS‐GFP or UAS‐HA‐dL(3)mbt;UAS‐GFP under vg‐Gal4 were reared to adulthood at 25°C. P‐values were estimated by comparing the proportions via a two‐proportion Z‐test. Scale bars: 200 μm.
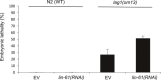
Functional interaction between lag‐1/RBPJ and lin‐61/L3MBTL3 during Caenorhabditis elegans vulva development. Proportion of animals (n ≥ 100) displaying a protruding vulva (Pvl) phenotype after RNAi treatment for two generations. Worms were grown at 25°C. Shown are means ± s.d. of duplicate experiments. EV, empty vector control.
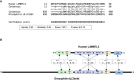
Summary of the analysis of the amino acid sequences of human L3MBTL3 and Drosophila dL(3)mbt using a hidden Markov model (HMM) profile alignment approach (Soding, 2005). L3MBTL3‐(11‐50) and dL(3)mbt‐(658‐698) regions are conserved (P = 6 × 10−19). The consensus sequences identified in the HMM profile–profile alignment analysis are series of tildes, and amino acid letters that represent the calculated order of most frequent residues found at each position in the multiple sequence alignment analyses for L3MBTL3 or dL(3)mbt and their homologs across species. An “uppercase letter” refers to a residue having high conservation in the profile. A “lowercase letter” refers to a residue having significant conservation in the profile. A “˜ symbol” refers to a position where no single residue stands out as being the most conserved. Between the alignments, the symbols indicate the overall value of aligning a pair of residues at a particular position: “.” indicates a score between −0.5 and +0.5; “+” indicates a score between +0.5 and +1.5; “|” indicates a score > +1.5; “empty space” indicates a gap in the alignment.
Schematic representation of the human L3MBTL3 and the Drosophila dL(3)mbt proteins. The analysis of the amino acid sequences using the HMM profile alignment approach generated highly confident alignments for seven conserved domains: the CSL interaction motif (CIM), the MBT domains #1, #2, and #3, the SAM domain, and the ZnF domains #1 and #2. P values are shown for each pair of conserved domains.
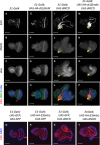
- A–D
Disks dissected from E1‐Gal4 control larvae present a normal morphology with a clear linear demarcation of EdU‐positive dividing cells at the level of the morphogenetic furrow (red arrow in panel D), an indentation that demarcates the boundary between elav‐positive developing photoreceptors located posteriorly and elav‐negative undifferentiated cells located anteriorly.
- E–H
HA‐dL(3)mbt overexpression alone has minimal effect on disk size or proliferation compared to E1‐Gal4 control.
- I–L
Ectopic expression of dNICD results in enlarged and distorted eye disks. Yellow arrows mark regions of high UAS‐dNICD expression.
- M–P
Gain of dL(3)mbt significantly suppresses the dNICD‐induced hyperplasia. Yellow arrows mark regions of high UAS‐dNICD expression.
- Q–T
To assess the potential effects associated with UAS titration, the number of UAS constructs was normalized with UAS‐GFP so that every genotype contained two UAS constructs. Disks were labeled with EdU (red) and counterstained with DAPI (blue). Note that the additional UAS‐GFP transgenes do not affect the EdU staining pattern or overall disk morphology (compare panels A, E, I, and M to panels Q, R, S, and T, respectively), demonstrating that UAS titration is not responsible for the UAS‐HA‐dL(3)mbt;UAS‐NICD phenotype.
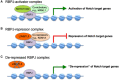
NOTCH ICD binds to RBPJ‐bound Notch‐responsive elements where it builds up a co‐activator complex composed of Mastermind‐like 1 (MAML1) and additional co‐activators to induce expression of Notch target genes.
In the absence of Notch signaling, L3MBTL3 interacts with RBPJ at Notch‐responsive elements where it recruits KDM1A to repress Notch target genes.
Loss of function of L3MBTL3 leads to de‐repression of Notch target genes.
Similar articles
-
The structure, binding and function of a Notch transcription complex involving RBPJ and the epigenetic reader protein L3MBTL3.Nucleic Acids Res. 2022 Dec 9;50(22):13083-13099. doi: 10.1093/nar/gkac1137. Nucleic Acids Res. 2022. PMID: 36477367 Free PMC article.
-
Nucleo-cytoplasmic shuttling of murine RBPJ by Hairless protein matches that of Su(H) protein in the model system Drosophila melanogaster.Hereditas. 2021 Mar 28;158(1):11. doi: 10.1186/s41065-021-00175-z. Hereditas. 2021. PMID: 33775255 Free PMC article.
-
Notch-independent functions of CSL.Curr Top Dev Biol. 2011;97:55-74. doi: 10.1016/B978-0-12-385975-4.00009-7. Curr Top Dev Biol. 2011. PMID: 22074602 Review.
-
Structural and functional analysis of the repressor complex in the Notch signaling pathway of Drosophila melanogaster.Mol Biol Cell. 2011 Sep;22(17):3242-52. doi: 10.1091/mbc.E11-05-0420. Epub 2011 Jul 7. Mol Biol Cell. 2011. PMID: 21737682 Free PMC article.
-
Transcription Factor RBPJ as a Molecular Switch in Regulating the Notch Response.Adv Exp Med Biol. 2021;1287:9-30. doi: 10.1007/978-3-030-55031-8_2. Adv Exp Med Biol. 2021. PMID: 33034023 Review.
Cited by
-
L3MBTL3 and STAT3 collaboratively upregulate SNAIL expression to promote metastasis in female breast cancer.Nat Commun. 2025 Jan 2;16(1):231. doi: 10.1038/s41467-024-55617-9. Nat Commun. 2025. PMID: 39747894 Free PMC article.
-
L3MBTL3 Is a Potential Prognostic Biomarker and Correlates with Immune Infiltrations in Gastric Cancer.Cancers (Basel). 2023 Dec 27;16(1):128. doi: 10.3390/cancers16010128. Cancers (Basel). 2023. PMID: 38201555 Free PMC article.
-
Uncovering immune cell-associated genes in breast cancer: based on summary data-based Mendelian randomized analysis and colocalization study.Breast Cancer Res. 2024 Nov 29;26(1):172. doi: 10.1186/s13058-024-01928-0. Breast Cancer Res. 2024. PMID: 39614330 Free PMC article.
-
Kir2.1 Interactome Mapping Uncovers PKP4 as a Modulator of the Kir2.1-Regulated Inward Rectifier Potassium Currents.Mol Cell Proteomics. 2020 Sep;19(9):1436-1449. doi: 10.1074/mcp.RA120.002071. Epub 2020 Jun 15. Mol Cell Proteomics. 2020. PMID: 32541000 Free PMC article.
-
Comprehensive genomic features indicative for Notch responsiveness.Nucleic Acids Res. 2024 May 22;52(9):5179-5194. doi: 10.1093/nar/gkae292. Nucleic Acids Res. 2024. PMID: 38647081 Free PMC article.
References
-
- Almeida MS, Bray SJ (2005) Regulation of post‐embryonic neuroblasts by Drosophila Grainyhead. Mech Dev 122: 1282–1293 - PubMed
-
- Amente S, Lania L, Majello B (2013) The histone LSD1 demethylase in stemness and cancer transcription programs. Biochim Biophys Acta 1829: 981–986 - PubMed
-
- Boccuni P, MacGrogan D, Scandura JM, Nimer SD (2003) The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J Biol Chem 278: 15412–15420 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

