BAD-LAMP controls TLR9 trafficking and signalling in human plasmacytoid dendritic cells
- PMID: 29030552
- PMCID: PMC5640662
- DOI: 10.1038/s41467-017-00695-1
BAD-LAMP controls TLR9 trafficking and signalling in human plasmacytoid dendritic cells
Abstract
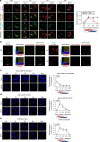
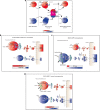
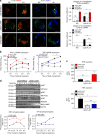
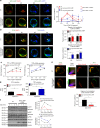
Toll-like receptors (TLR) are essential components of the innate immune system. Several accessory proteins, such as UNC93B1, are required for transport and activation of nucleic acid sensing Toll-like receptors in endosomes. Here, we show that BAD-LAMP (LAMP5) controls TLR9 trafficking to LAMP1+ late endosomes in human plasmacytoid dendritic cells (pDC), leading to NF-κB activation and TNF production upon DNA detection. An inducible VAMP3+/LAMP2+/LAMP1- endolysosome compartment exists in pDCs from which TLR9 activation triggers type I interferon expression. BAD-LAMP-silencing enhances TLR9 retention in this compartment and consequent downstream signalling events. Conversely, sustained BAD-LAMP expression in pDCs contributes to their lack of type I interferon production after exposure to a TGF-β-positive microenvironment or isolation from human breast tumours. Hence, BAD-LAMP limits interferon expression in pDCs indirectly, by promoting TLR9 sorting to late endosome compartments at steady state and in response to immunomodulatory cues.TLR9 is highly expressed by plasmacytoid dendritic cells and detects nucleic acids, but to discriminate between host and microbial nucleic acids TLR9 is sorted into different endosomal compartments. Here the authors show that BAD-LAMP limits type 1 interferon responses by sorting TLR9 to late endosomal compartments.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

