Ribosome Profiling Reveals Genome-wide Cellular Translational Regulation upon Heat Stress in Escherichia coli
- PMID: 29031842
- PMCID: PMC5673677
- DOI: 10.1016/j.gpb.2017.04.005
Ribosome Profiling Reveals Genome-wide Cellular Translational Regulation upon Heat Stress in Escherichia coli
Abstract
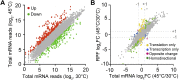
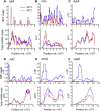
Heat shock response is a classical stress-induced regulatory system in bacteria, characterized by extensive transcriptional reprogramming. To compare the impact of heat stress on the transcriptome and translatome in Escherichia coli, we conducted ribosome profiling in parallel with RNA-Seq to investigate the alterations in transcription and translation efficiency when E. coli cells were exposed to a mild heat stress (from 30 °C to 45 °C). While general changes in ribosome footprints correlate with the changes of mRNA transcripts upon heat stress, a number of genes show differential changes at the transcription and translation levels. Translation efficiency of a few genes that are related to environment stimulus response is up-regulated, and in contrast, some genes functioning in mRNA translation and amino acid biosynthesis are down-regulated at the translation level in response to heat stress. Moreover, our ribosome occupancy data suggest that in general ribosomes accumulate remarkably in the starting regions of ORFs upon heat stress. This study provides additional insights into bacterial gene expression in response to heat stress, and suggests the presence of stress-induced but yet-to-be characterized cellular regulatory mechanisms of gene expression at translation level.
Keywords: Heat shock response; RNA-Seq; Ribosome profiling; Transcription regulation; Translation regulation.
Copyright © 2017 The Authors. Production and hosting by Elsevier B.V. All rights reserved.
Figures

References
-
- Arsene F., Tomoyasu T., Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. - PubMed
-
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. - PubMed
-
- Wang Z.X., Zhou X.Z., Meng H.M., Liu Y.J., Zhou Q., Huang B. Comparative transcriptomic analysis of the heat stress response in the filamentous fungus Metarhizium anisopliae using RNA-Seq. Appl Microbiol Biotechnol. 2014;98:5589–5597. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

