Nanodisc-based kinetic assays reveal distinct effects of phospholipid headgroups on the phosphoenzyme transition of sarcoplasmic reticulum Ca2+-ATPase
- PMID: 29032359
- PMCID: PMC5724008
- DOI: 10.1074/jbc.M117.816702
Nanodisc-based kinetic assays reveal distinct effects of phospholipid headgroups on the phosphoenzyme transition of sarcoplasmic reticulum Ca2+-ATPase
Abstract
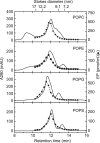
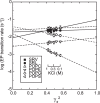
Sarco(endo)plasmic reticulum Ca2+-ATPase catalyzes ATP-driven Ca2+ transport from the cytoplasm to the lumen and is critical for a range of cell functions, including muscle relaxation. Here, we investigated the effects of the headgroups of the 1-palmitoyl-2-oleoyl glycerophospholipids phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylglycerol (PG) on sarcoplasmic reticulum (SR) Ca2+-ATPase embedded into a nanodisc, a lipid-bilayer construct harboring the specific lipid. We found that Ca2+-ATPase activity in a PC bilayer is comparable with that of SR vesicles and is suppressed in the other phospholipids, especially in PS. Ca2+ affinity at the high-affinity transport sites in PC was similar to that of SR vesicles, but 2-3-fold reduced in PE and PS. Ca2+ on- and off-rates in the non-phosphorylated ATPase were markedly reduced in PS. Rate-limiting phosphoenzyme (EP) conformational transition in 0.1 m KCl was as rapid in PC as in SR vesicles, but slowed in other phospholipids, especially in PS. Using kinetic plots of the logarithm of rate versus the square of mean activity coefficient of solutes in 0.1-1 m KCl, we noted that PC is optimal for the EP transition, but PG and especially PS had markedly unfavorable electrostatic effects, and PE exhibited a strong non-electrostatic restriction. Thus, the major SR membrane lipid PC is optimal for all steps and, unlike the other headgroups, contributes favorable electrostatics and non-electrostatic elements during the EP transition. Our analyses further revealed that the surface charge of the lipid bilayer directly modulates the transition rate.
Keywords: calcium ATPase; kinetics; lipid-protein interaction; membrane enzyme; phospholipid; sarcoplasmic reticulum (SR).
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
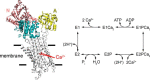






References
-
- Toyoshima C. (2008) Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch. Biochem. Biophys. 476, 3–11 - PubMed
-
- Toyoshima C. (2009) How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta 1793, 941–946 - PubMed
-
- Møller J. V., Olesen C., Winther A.-M. L., and Nissen P. (2010) The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q. Rev. Biophys. 43, 501–566 - PubMed
-
- Toyoshima C., Nakasako M., Nomura H., and Ogawa H. (2000) Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 - PubMed
-
- Danko S., Yamasaki K., Daiho T., and Suzuki H., and Toyoshima C. (2001) Organization of cytoplasmic domains of sarcoplasmic reticulum Ca2+-ATPase in E1P and E1ATP states: a limited proteolysis study. FEBS Lett. 505, 129–135 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

