Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction
- PMID: 29033129
- PMCID: PMC5683101
- DOI: 10.1016/j.cell.2017.09.018
Genome-Nuclear Lamina Interactions Regulate Cardiac Stem Cell Lineage Restriction
Abstract
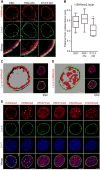
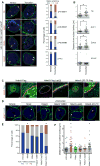
Progenitor cells differentiate into specialized cell types through coordinated expression of lineage-specific genes and modification of complex chromatin configurations. We demonstrate that a histone deacetylase (Hdac3) organizes heterochromatin at the nuclear lamina during cardiac progenitor lineage restriction. Specification of cardiomyocytes is associated with reorganization of peripheral heterochromatin, and independent of deacetylase activity, Hdac3 tethers peripheral heterochromatin containing lineage-relevant genes to the nuclear lamina. Deletion of Hdac3 in cardiac progenitor cells releases genomic regions from the nuclear periphery, leading to precocious cardiac gene expression and differentiation into cardiomyocytes; in contrast, restricting Hdac3 to the nuclear periphery rescues myogenesis in progenitors otherwise lacking Hdac3. Our results suggest that availability of genomic regions for activation by lineage-specific factors is regulated in part through dynamic chromatin-nuclear lamina interactions and that competence of a progenitor cell to respond to differentiation signals may depend upon coordinated movement of responding gene loci away from the nuclear periphery.
Keywords: cardiac specification; cellular competence; genome organization; lineage restriction; nuclear lamina.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures





References
-
- Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, Cerase A, McDonel P, Guttman M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. - PubMed
-
- Filion GJ, van Steensel B. Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells. Nat Genet. 2010;42:4. author reply 5–6. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

