A pUL25 dimer interfaces the pseudorabies virus capsid and tegument
- PMID: 29035172
- PMCID: PMC5718256
- DOI: 10.1099/jgv.0.000903
A pUL25 dimer interfaces the pseudorabies virus capsid and tegument
Abstract
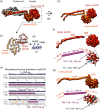
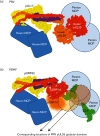
Inside the virions of α-herpesviruses, tegument protein pUL25 anchors the tegument to capsid vertices through direct interactions with tegument proteins pUL17 and pUL36. In addition to promoting virion assembly, both pUL25 and pUL36 are critical for intracellular microtubule-dependent capsid transport. Despite these essential roles during infection, the stoichiometry and precise organization of pUL25 and pUL36 on the capsid surface remain controversial due to the insufficient resolution of existing reconstructions from cryo-electron microscopy (cryoEM). Here, we report a three-dimensional (3D) icosahedral reconstruction of pseudorabies virus (PRV), a varicellovirus of the α-herpesvirinae subfamily, obtained by electron-counting cryoEM at 4.9 Å resolution. Our reconstruction resolves a dimer of pUL25 forming a capsid-associated tegument complex with pUL36 and pUL17 through a coiled coil helix bundle, thus correcting previous misinterpretations. A comparison between reconstructions of PRV and the γ-herpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV) reinforces their similar architectures and establishes important subfamily differences in the capsid-tegument interface.
Keywords: CryoEM; pUL17; pUL25 dimer; pUL36 (VP1/2); pseudorabies virus; tegument proteins.
Figures




Similar articles
-
Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins.J Mol Biol. 2013 Sep 23;425(18):3415-28. doi: 10.1016/j.jmb.2013.06.034. Epub 2013 Jul 1. J Mol Biol. 2013. PMID: 23827137 Free PMC article.
-
The large tegument protein pUL36 is essential for formation of the capsid vertex-specific component at the capsid-tegument interface of herpes simplex virus 1.J Virol. 2015 Feb;89(3):1502-11. doi: 10.1128/JVI.02887-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410861 Free PMC article.
-
Organization of capsid-associated tegument components in Kaposi's sarcoma-associated herpesvirus.J Virol. 2014 Nov;88(21):12694-702. doi: 10.1128/JVI.01509-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142590 Free PMC article.
-
Herpesvirus Capsid Assembly and DNA Packaging.Adv Anat Embryol Cell Biol. 2017;223:119-142. doi: 10.1007/978-3-319-53168-7_6. Adv Anat Embryol Cell Biol. 2017. PMID: 28528442 Free PMC article. Review.
-
Comprehensive Analysis of the Tegument Proteins Involved in Capsid Transport and Virion Morphogenesis of Alpha, Beta and Gamma Herpesviruses.Viruses. 2023 Oct 6;15(10):2058. doi: 10.3390/v15102058. Viruses. 2023. PMID: 37896835 Free PMC article. Review.
Cited by
-
Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.Nat Microbiol. 2020 Oct;5(10):1285-1298. doi: 10.1038/s41564-020-0758-1. Epub 2020 Jul 27. Nat Microbiol. 2020. PMID: 32719506 Free PMC article.
-
DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.Cell. 2019 Sep 5;178(6):1329-1343.e12. doi: 10.1016/j.cell.2019.07.035. Epub 2019 Aug 22. Cell. 2019. PMID: 31447177 Free PMC article.
-
Herpesvirus acts with the cytoskeleton and promotes cancer progression.J Cancer. 2019 May 21;10(10):2185-2193. doi: 10.7150/jca.30222. eCollection 2019. J Cancer. 2019. PMID: 31258722 Free PMC article. Review.
-
Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150.Nat Commun. 2021 Sep 17;12(1):5513. doi: 10.1038/s41467-021-25791-1. Nat Commun. 2021. PMID: 34535641 Free PMC article.
-
Bovine Herpesvirus 1 Invasion of Sensory Neurons by Retrograde Axonal Transport Is Dependent on the pUL37 Region 2 Effector.J Virol. 2022 May 11;96(9):e0148621. doi: 10.1128/jvi.01486-21. Epub 2022 Apr 14. J Virol. 2022. PMID: 35420461 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

