PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types
- PMID: 29036324
- PMCID: PMC5753226
- DOI: 10.1093/nar/gkx861
PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types
Abstract
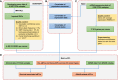
Expression quantitative trait locus (eQTL) analysis, which links variations in gene expression to genotypes, is essential to understanding gene regulation and to interpreting disease-associated loci. Currently identified eQTLs are mainly in samples of blood and other normal tissues. However, no database comprehensively provides eQTLs in large number of cancer samples. Using the genotype and expression data of 9196 tumor samples in 33 cancer types from The Cancer Genome Atlas (TCGA), we identified 5 606 570 eQTL-gene pairs in the cis-eQTL analysis and 231 210 eQTL-gene pairs in the trans-eQTL analysis. We further performed survival analysis and identified 22 212 eQTLs associated with patient overall survival. Furthermore, we linked the eQTLs to genome-wide association studies (GWAS) data and identified 337 131 eQTLs that overlap with existing GWAS loci. We developed PancanQTL, a user-friendly database (http://bioinfo.life.hust.edu.cn/PancanQTL/), to store cis-eQTLs, trans-eQTLs, survival-associated eQTLs and GWAS-related eQTLs to enable searching, browsing and downloading. PancanQTL could help the research community understand the effects of inherited variants in tumorigenesis and development.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Wu C., Miao X., Huang L., Che X., Jiang G., Yu D., Yang X., Cao G., Hu Z., Zhou Y. et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat. Genet. 2011; 44:62–66. - PubMed
-
- Schork N.J., Fallin D., Lanchbury J.S.. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin. Genet. 2000; 58:250–264. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

