Transcription-Associated Compositional Skews in Drosophila Genes
- PMID: 29036491
- PMCID: PMC5786239
- DOI: 10.1093/gbe/evx200
Transcription-Associated Compositional Skews in Drosophila Genes
Abstract
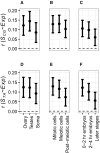
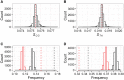
In many organisms, local deviations from Chargaff's second parity rule are observed around replication and transcription start sites and within intron sequences. Here, we use expression data as well as a whole-genome data set of nearly 200 haplotypes to investigate such compositional skews in Drosophila melanogaster genes. We find a positive correlation between compositional skew and gene expression, comparable in strength to similar correlations between expression levels and genome-wide sequence features. This correlation is relatively stronger for germline, compared with somatic expression, consistent with the process of transcription-associated mutation bias. We also inferred mutation rates from alleles segregating at low frequencies in short introns, and show that, whereas the overall GC content of short introns does not conform to the equilibrium expectation, the level of the observed deviation from the second parity rule is generally consistent with the inferred rates.
Keywords: Chargaff’s second parity rule; base composition evolution; compositional skew; transcription-associated mutation bias.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Andolfatto P. 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 4377062:1149–1152. - PubMed
-
- Arneodo A, et al. 2011. Multi-scale coding of genomic information: from DNA sequence to genome structure and function. Phys Rep. 498(2-3):45–188.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

