Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications
- PMID: 29036500
- PMCID: PMC5892155
- DOI: 10.1093/neuonc/nox183
Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications
Abstract
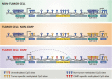
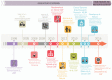
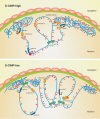
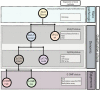
Gliomas are a heterogeneous group of brain tumors with distinct biological and clinical properties. Despite advances in surgical techniques and clinical regimens, treatment of high-grade glioma remains challenging and carries dismal rates of therapeutic success and overall survival. Challenges include the molecular complexity of gliomas, as well as inconsistencies in histopathological grading, resulting in an inaccurate prediction of disease progression and failure in the use of standard therapy. The updated 2016 World Health Organization (WHO) classification of tumors of the central nervous system reflects a refinement of tumor diagnostics by integrating the genotypic and phenotypic features, thereby narrowing the defined subgroups. The new classification recommends molecular diagnosis of isocitrate dehydrogenase (IDH) mutational status in gliomas. IDH-mutant gliomas manifest the cytosine-phosphate-guanine (CpG) island methylator phenotype (G-CIMP). Notably, the recent identification of clinically relevant subsets of G-CIMP tumors (G-CIMP-high and G-CIMP-low) provides a further refinement in glioma classification that is independent of grade and histology. This scheme may be useful for predicting patient outcome and may be translated into effective therapeutic strategies tailored to each patient. In this review, we highlight the evolution of our understanding of the G-CIMP subsets and how recent advances in characterizing the genome and epigenome of gliomas may influence future basic and translational research.
Figures

References
-
- Louis DN, Perry A, Reifenberger G et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. - PubMed
-
- Masui K, Mischel PS, Reifenberger G. Molecular classification of gliomas. Handb Clin Neurol. 2016;134:97–120. - PubMed
-
- Siegal T. Clinical relevance of prognostic and predictive molecular markers in gliomas. Adv Tech Stand Neurosurg 2016;(43):91–108. - PubMed
-
- Taylor JW, Chi AS, Cahill DP. Tailored therapy in diffuse gliomas: using molecular classifiers to optimize clinical management. Oncology (Williston Park). 2013;27(6):504–514. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

