ComplexViewer: visualization of curated macromolecular complexes
- PMID: 29036573
- PMCID: PMC5870653
- DOI: 10.1093/bioinformatics/btx497
ComplexViewer: visualization of curated macromolecular complexes
Abstract
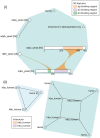
Summary: Proteins frequently function as parts of complexes, assemblages of multiple proteins and other biomolecules, yet network visualizations usually only show proteins as parts of binary interactions. ComplexViewer visualizes interactions with more than two participants and thereby avoids the need to first expand these into multiple binary interactions. Furthermore, if binding regions between molecules are known then these can be displayed in the context of the larger complex.
Availability and implementation: freely available under Apache version 2 license; EMBL-EBI Complex Portal: http://www.ebi.ac.uk/complexportal; Source code: https://github.com/MICommunity/ComplexViewer; Package: https://www.npmjs.com/package/complexviewer; http://biojs.io/d/complexviewer. Language: JavaScript; Web technology: Scalable Vector Graphics; Libraries: D3.js.
Contact: colin.combe@ed.ac.uk or juri.rappsilber@ed.ac.uk.
© The Author 2017. Published by Oxford University Press.
Figures
References
-
- Hermjakob H. et al. (2004) The HUPO PSI’s Molecular Interaction format—a community standard for the representation of protein interaction data. Nat. Biotechnol., 22, 177–183. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

