The effects of DNA supercoiling on G-quadruplex formation
- PMID: 29036619
- PMCID: PMC5716088
- DOI: 10.1093/nar/gkx856
The effects of DNA supercoiling on G-quadruplex formation
Abstract
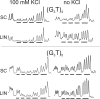
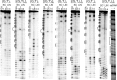
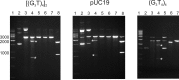
Guanine-rich DNAs can fold into four-stranded structures that contain stacks of G-quartets. Bioinformatics studies have revealed that G-rich sequences with the potential to adopt these structures are unevenly distributed throughout genomes, and are especially found in gene promoter regions. With the exception of the single-stranded telomeric DNA, all genomic G-rich sequences will always be present along with their C-rich complements, and quadruplex formation will be in competition with the corresponding Watson-Crick duplex. Quadruplex formation must therefore first require local dissociation (melting) of the duplex strands. Since negative supercoiling is known to facilitate the formation of alternative DNA structures, we have investigated G-quadruplex formation within negatively supercoiled DNA plasmids. Plasmids containing multiple copies of (G3T)n and (G3T4)n repeats, were probed with dimethylsulphate, potassium permanganate and S1 nuclease. While dimethylsulphate footprinting revealed some evidence for G-quadruplex formation in (G3T)n sequences, this was not affected by supercoiling, and permanganate failed to detect exposed thymines in the loop regions. (G3T4)n sequences were not protected from DMS and showed no reaction with permanganate. Similarly, both S1 nuclease and 2D gel electrophoresis of DNA topoisomers did not detect any supercoil-dependent structural transitions. These results suggest that negative supercoiling alone is not sufficient to drive G-quadruplex formation.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


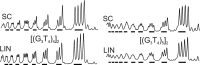
Similar articles
-
Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents.Nucleic Acids Res. 2005 Oct 20;33(18):6070-80. doi: 10.1093/nar/gki917. Print 2005. Nucleic Acids Res. 2005. PMID: 16239639 Free PMC article.
-
Solution equilibria of cytosine- and guanine-rich sequences near the promoter region of the n-myc gene that contain stable hairpins within lateral loops.Biochim Biophys Acta. 2014 Jan;1840(1):41-52. doi: 10.1016/j.bbagen.2013.08.028. Epub 2013 Sep 6. Biochim Biophys Acta. 2014. PMID: 24012973
-
G-quadruplex formation between G-rich PNA and homologous sequences in oligonucleotides and supercoiled plasmid DNA.Nucleic Acid Ther. 2015 Apr;25(2):78-84. doi: 10.1089/nat.2014.0517. Epub 2015 Feb 4. Nucleic Acid Ther. 2015. PMID: 25650982 Free PMC article.
-
Methods for investigating G-quadruplex DNA/ligand interactions.Chem Soc Rev. 2011 Nov;40(11):5293-307. doi: 10.1039/c1cs15117g. Epub 2011 Jul 1. Chem Soc Rev. 2011. PMID: 21720638 Review.
-
Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids.Biopolymers. 2000-2001;56(3):147-94. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. Biopolymers. 2000. PMID: 11745110 Review.
Cited by
-
Crosstalk between G-Quadruplexes and Dnmt3a-Mediated Methylation of the c-MYC Oncogene Promoter.Int J Mol Sci. 2023 Dec 19;25(1):45. doi: 10.3390/ijms25010045. Int J Mol Sci. 2023. PMID: 38203216 Free PMC article.
-
Aclarubicin stimulates RNA polymerase II elongation at closely spaced divergent promoters.Sci Adv. 2023 Jun 16;9(24):eadg3257. doi: 10.1126/sciadv.adg3257. Epub 2023 Jun 14. Sci Adv. 2023. PMID: 37315134 Free PMC article.
-
Insight into formation propensity of pseudocircular DNA G-hairpins.Nucleic Acids Res. 2021 Feb 26;49(4):2317-2332. doi: 10.1093/nar/gkab029. Nucleic Acids Res. 2021. PMID: 33524154 Free PMC article.
-
Creation and resolution of non-B-DNA structural impediments during replication.Crit Rev Biochem Mol Biol. 2022 Aug;57(4):412-442. doi: 10.1080/10409238.2022.2121803. Epub 2022 Sep 28. Crit Rev Biochem Mol Biol. 2022. PMID: 36170051 Free PMC article.
-
Design and Properties of Ligand-Conjugated Guanine Oligonucleotides for Recovery of Mutated G-Quadruplexes.Molecules. 2018 Dec 6;23(12):3228. doi: 10.3390/molecules23123228. Molecules. 2018. PMID: 30563296 Free PMC article.
References
-
- Sen D., Gilbert W.. Formation of parallel 4-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. - PubMed
-
- Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol. Chem. 2001; 382:621–628. - PubMed
-
- Huppert J.L. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008; 37:1375–1384. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

