Trypanosoma cruzi specific mRNA amplification by in vitro transcription improves parasite transcriptomics in host-parasite RNA mixtures
- PMID: 29037144
- PMCID: PMC5644099
- DOI: 10.1186/s12864-017-4163-y
Trypanosoma cruzi specific mRNA amplification by in vitro transcription improves parasite transcriptomics in host-parasite RNA mixtures
Abstract
Background: Trypanosomatids are a group of protozoan parasites that includes the etiologic agents of important human illnesses as Chagas disease, sleeping sickness and leishmaniasis. These parasites have a significant distinction from other eukaryotes concerning mRNA structure, since all mature mRNAs have an identical species-specific sequence of 39 nucleotides at the 5' extremity, named spliced leader (SL). Considering this peculiar aspect of trypanosomatid mRNA, the aim of the present work was to develop a Trypanosoma cruzi specific in vitro transcription (IVT) linear mRNA amplification method in order to improve parasite transcriptomics analyses.
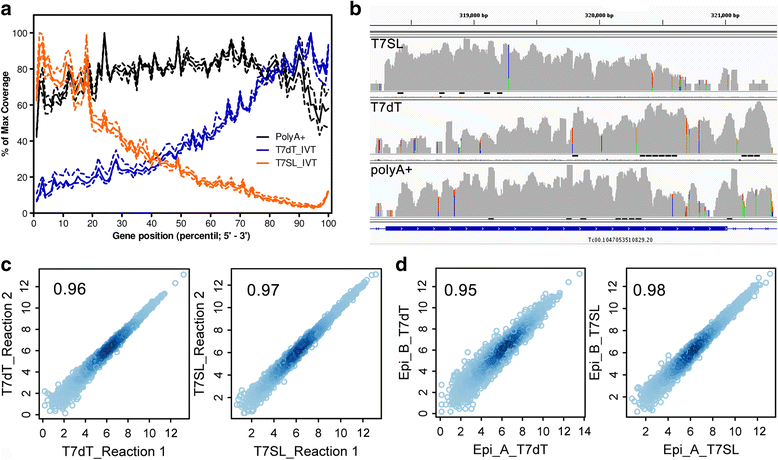
Methods: We designed an oligonucleotide complementary to the last 21 bases of T. cruzi SL sequence, bearing an upstream T7 promoter (T7SL primer), which was used to direct the synthesis of second-strand cDNA. Original mRNA was then amplified by IVT using T7 RNA polymerase. T7SL-amplified RNA from two distinct T. cruzi stages (epimastigotes and trypomastigotes) were deep sequenced in SOLiD platform. Usual poly(A) + RNA and and T7-oligo(dT) amplified RNA (Eberwine method) were also sequenced. RNA-Seq reads were aligned to our new and improved T. cruzi Dm28c genome assembly (PacBio technology) and resulting transcriptome pattern from these three RNA preparation methods were compared, mainly concerning the conservation of mRNA transcritional levels and DEGs detection between epimastigotes and trypomastigotes.
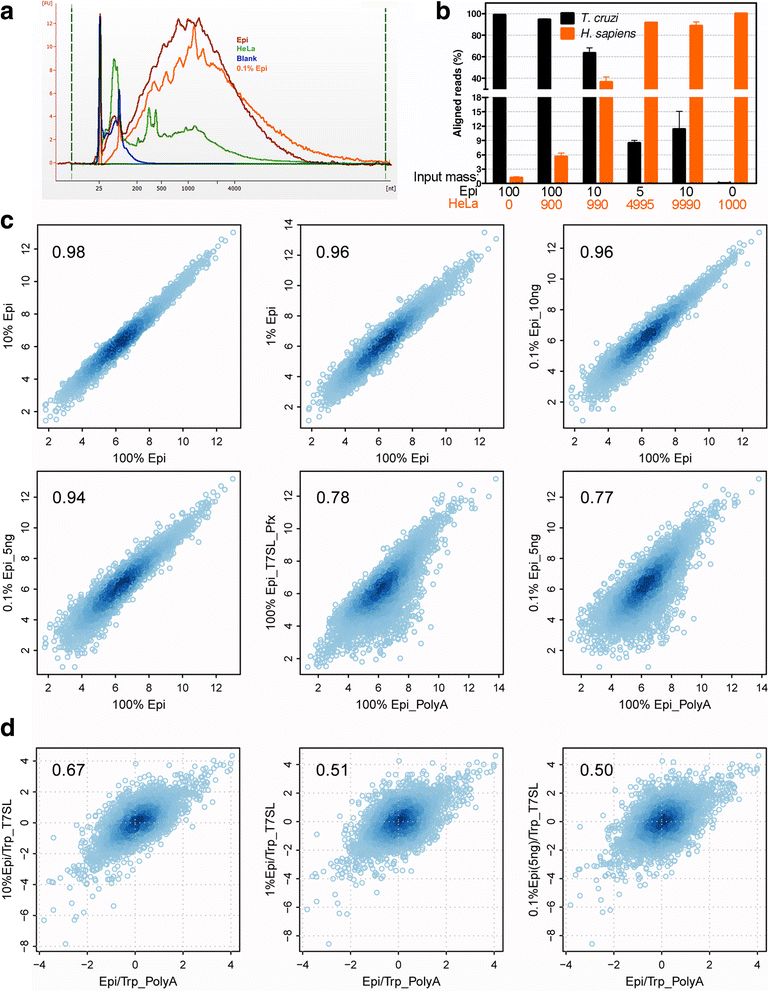
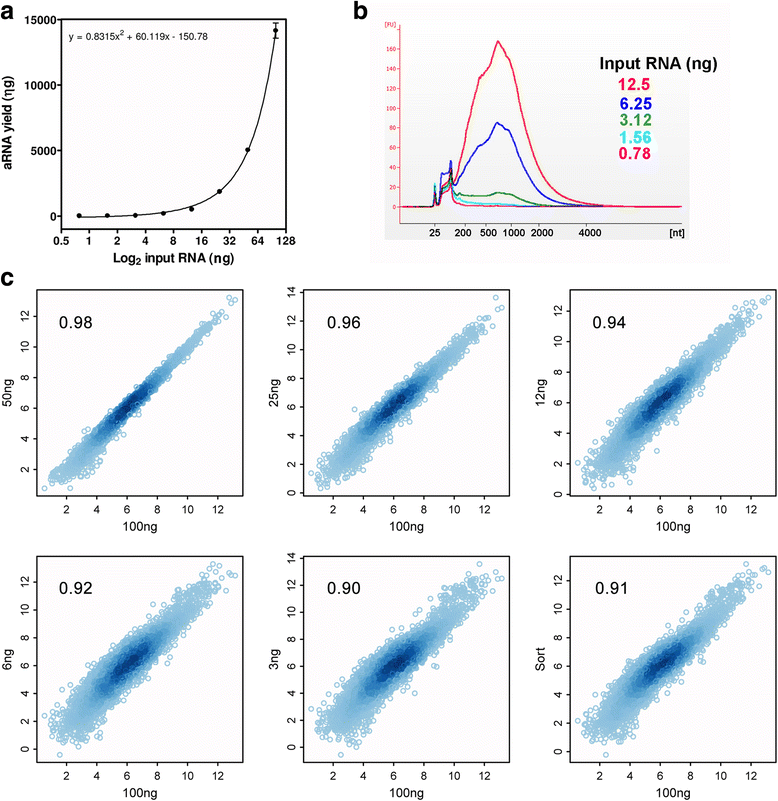
Results: T7SL IVT method detected more potential differentially expressed genes in comparison to either poly(A) + RNA or T7dT IVT, and was also able to produce reliable quantifications of the parasite transcriptome down to 3 ng of total RNA. Furthermore, amplification of parasite mRNA in HeLa/epimastigote RNA mixtures showed that T7SL IVT generates transcriptome quantification with similar detection of differentially expressed genes when parasite RNA mass was only 0.1% of the total mixture (R = 0.78 when compared to poly(A) + RNA).
Conclusions: The T7SL IVT amplification method presented here allows the detection of more potential parasite differentially expressed genes (in comparison to poly(A) + RNA) in host-parasite mixtures or samples with low amount of RNA. This method is especially useful for trypanosomatid transcriptomics because it produces less bias than PCR-based mRNA amplification. Additionally, by simply changing the complementary region of the T7SL primer, the present method can be applied to any trypanosomatid species.
Keywords: RNA amplification; RNA-Seq; T7 RNA polymerase; Transcriptomics; Trypanosoma cruzi; Trypanosomatid.
Conflict of interest statement
Competing interest
The authors declare that they have no competing interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

