Generalized disequilibrium test for association in qualitative traits incorporating imprinting effects based on extended pedigrees
- PMID: 29037145
- PMCID: PMC5644153
- DOI: 10.1186/s12863-017-0560-0
Generalized disequilibrium test for association in qualitative traits incorporating imprinting effects based on extended pedigrees
Abstract
Background: For dichotomous traits, the generalized disequilibrium test with the moment estimate of the variance (GDT-ME) is a powerful family-based association method. Genomic imprinting is an important epigenetic phenomenon and currently, there has been increasing interest of incorporating imprinting to improve the test power of association analysis. However, GDT-ME does not take imprinting effects into account, and it has not been investigated whether it can be used for association analysis when the effects indeed exist.
Results: In this article, based on a novel decomposition of the genotype score according to the paternal or maternal source of the allele, we propose the generalized disequilibrium test with imprinting (GDTI) for complete pedigrees without any missing genotypes. Then, we extend GDTI and GDT-ME to accommodate incomplete pedigrees with some pedigrees having missing genotypes, by using a Monte Carlo (MC) sampling and estimation scheme to infer missing genotypes given available genotypes in each pedigree, denoted by MCGDTI and MCGDT-ME, respectively. The proposed GDTI and MCGDTI methods evaluate the differences of the paternal as well as maternal allele scores for all discordant relative pairs in a pedigree, including beyond first-degree relative pairs. Advantages of the proposed GDTI and MCGDTI test statistics over existing methods are demonstrated by simulation studies under various simulation settings and by application to the rheumatoid arthritis dataset. Simulation results show that the proposed tests control the size well under the null hypothesis of no association, and outperform the existing methods under various imprinting effect models. The existing GDT-ME and the proposed MCGDT-ME can be used to test for association even when imprinting effects exist. For the application to the rheumatoid arthritis data, compared to the existing methods, MCGDTI identifies more loci statistically significantly associated with the disease.
Conclusions: Under complete and incomplete imprinting effect models, our proposed GDTI and MCGDTI methods, by considering the information on imprinting effects and all discordant relative pairs within each pedigree, outperform all the existing test statistics and MCGDTI can recapture much of the missing information. Therefore, MCGDTI is recommended in practice.
Keywords: Generalized disequilibrium test; Genomic imprinting; Monte Carlo sampling; Qualitative trait.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Figures

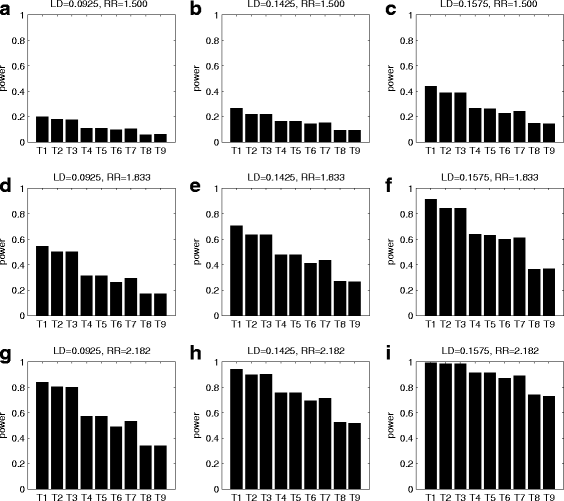
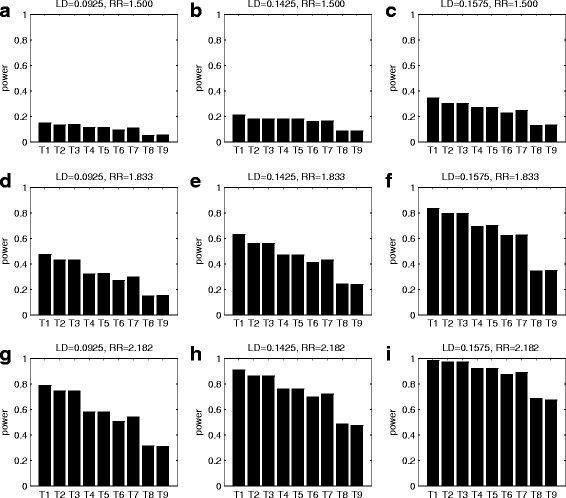

Similar articles
-
A powerful association test for qualitative traits incorporating imprinting effects using general pedigree data.J Hum Genet. 2015 Feb;60(2):77-83. doi: 10.1038/jhg.2014.109. Epub 2014 Dec 18. J Hum Genet. 2015. PMID: 25518739
-
A powerful parent-of-origin effects test for qualitative traits on X chromosome in general pedigrees.BMC Bioinformatics. 2018 Jan 5;19(1):8. doi: 10.1186/s12859-017-2001-5. BMC Bioinformatics. 2018. PMID: 29304743 Free PMC article.
-
The Rare-Variant Generalized Disequilibrium Test for Association Analysis of Nuclear and Extended Pedigrees with Application to Alzheimer Disease WGS Data.Am J Hum Genet. 2017 Feb 2;100(2):193-204. doi: 10.1016/j.ajhg.2016.12.001. Epub 2017 Jan 5. Am J Hum Genet. 2017. PMID: 28065470 Free PMC article.
-
QTL analysis in arbitrary pedigrees with incomplete marker information.Heredity (Edinb). 2002 Nov;89(5):339-45. doi: 10.1038/sj.hdy.6800136. Heredity (Edinb). 2002. PMID: 12399991 Review.
-
Linkage analysis with sequential imputation.Genet Epidemiol. 2003 Jul;25(1):25-35. doi: 10.1002/gepi.10249. Genet Epidemiol. 2003. PMID: 12813724 Review.
References
-
- Morison IM, Paton CJ, Cleverley SD. The imprinted gene and parent-of-origin effect database. 2001. http://igc.otago.ac.nz. Accessed 26 Mar 2017. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

