Neuroepigenetic mechanisms in disease
- PMID: 29037228
- PMCID: PMC5644115
- DOI: 10.1186/s13072-017-0150-4
Neuroepigenetic mechanisms in disease
Abstract
Epigenetics allows for the inheritance of information in cellular lineages during differentiation, independent of changes to the underlying genetic sequence. This raises the question of whether epigenetic mechanisms also function in post-mitotic neurons. During the long life of the neuron, fluctuations in gene expression allow the cell to pass through stages of differentiation, modulate synaptic activity in response to environmental cues, and fortify the cell through age-related neuroprotective pathways. Emerging evidence suggests that epigenetic mechanisms such as DNA methylation and histone modification permit these dynamic changes in gene expression throughout the life of a neuron. Accordingly, recent studies have revealed the vital importance of epigenetic players in the central nervous system and during neurodegeneration. Here, we provide a review of several of these recent findings, highlighting novel functions for epigenetics in the fields of Rett syndrome, Fragile X syndrome, and Alzheimer's disease research. Together, these discoveries underscore the vital importance of epigenetics in human neurological disorders.
Keywords: Alzheimer’s disease; DNA methylation; FMR1; Fragile X syndrome; Histone modification; LSD1/KDM1A; MECP2; Neuroepigenetics; Rett syndrome.
Figures
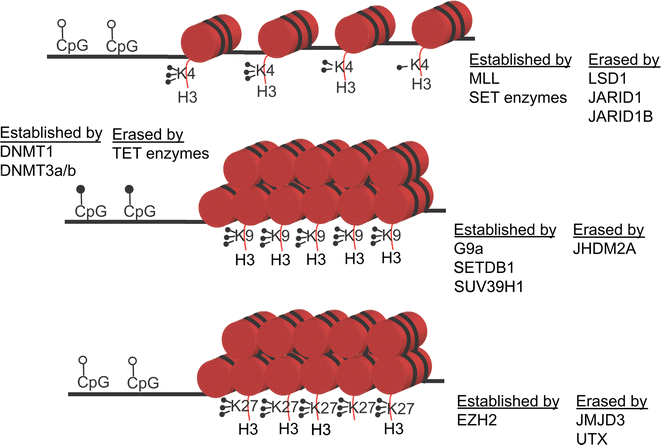
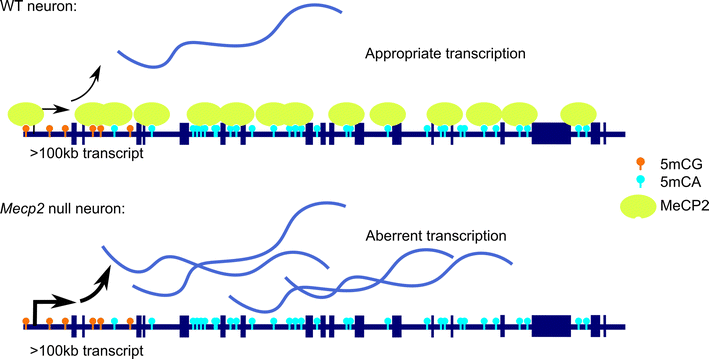
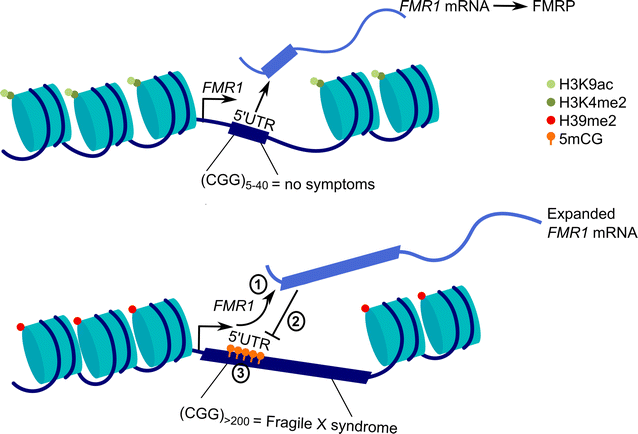

References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. - PubMed
-
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. - PubMed
-
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

