Co-Localization of Insulin-Like Growth Factor Binding Protein-1, Casein Kinase-2β, and Mechanistic Target of Rapamycin in Human Hepatocellular Carcinoma Cells as Demonstrated by Dual Immunofluorescence and in Situ Proximity Ligation Assay
- PMID: 29037858
- PMCID: PMC5745526
- DOI: 10.1016/j.ajpath.2017.09.009
Co-Localization of Insulin-Like Growth Factor Binding Protein-1, Casein Kinase-2β, and Mechanistic Target of Rapamycin in Human Hepatocellular Carcinoma Cells as Demonstrated by Dual Immunofluorescence and in Situ Proximity Ligation Assay
Abstract
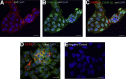
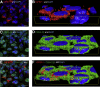
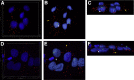
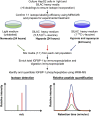
Insulin-like growth factor binding protein (IGFBP)-1 influences fetal growth by modifying insulin-like growth factor-I (IGF-I) bioavailability. IGFBP-1 phosphorylation, which markedly increases its affinity for IGF-I, is regulated by mechanistic target of rapamycin (mTOR) and casein kinase (CSNK)-2. However, the underlying molecular mechanisms remain unknown. We examined the cellular localization and potential interactions of IGFBP-1, CSNK-2β, and mTOR as a prerequisite for protein-protein interaction. Analysis of dual immunofluorescence images indicated a potential perinuclear co-localization between IGFBP-1 and CSNK-2β and a nuclear co-localization between CSNK-2β and mTOR. Proximity ligation assay (PLA) indicated proximity between IGFBP-1 and CSNK-2β as well as mTOR and CSNK-2β but not between mTOR and IGFBP-1. Three-dimensional rendering of the PLA images validated that IGFBP-1 and CSNK-2β interactions were in the perinuclear region and mTOR and CSNK-2β interactions were also predominantly perinuclear rather than nuclear as indicated by mTOR and CSNK-2β co-localization. Compared with control, hypoxia and rapamycin treatment showed markedly amplified PLA signals for IGFBP-1 and CSNK-2β (approximately 18-fold, P = 0.0002). Stable isotope labeling with multiple reaction monitoring-mass spectrometry demonstrated that hypoxia and rapamycin treatment increased IGFBP-1 phosphorylation at Ser98/Ser101/Ser119/Ser174 but most considerably (106-fold) at Ser169. We report interactions between CSNK-2β and IGFBP-1 as well as mTOR and CSNK-2β, providing strong evidence of a mechanistic link between mTOR and IGF-I signaling, two critical regulators of cell growth via CSNK-2.
Copyright © 2018 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Han V.K., Matsell D.G., Delhanty P.J., Hill D.J., Shimasaki S., Nygard K. IGF-binding protein mRNAs in the human fetus: tissue and cellular distribution of developmental expression. Horm Res. 1996;45:160–166. - PubMed
-
- Jones J.I., Busby W.H., Jr., Wright G., Clemmons D.R. Human IGFBP-1 is phosphorylated on 3 serine residues: effects of site-directed mutagenesis of the major phosphoserine. Growth Regul. 1993;3:37–40. - PubMed
-
- Martina N.A., Kim E., Chitkara U., Wathen N.C., Chard T., Giudice L.C. Gestational age-dependent expression of insulin-like growth factor-binding protein-1 (IGFBP-1) phosphoisoforms in human extraembryonic cavities, maternal serum, and decidua suggests decidua as the primary source of IGFBP-1 in these fluids during early pregnancy. J Clin Endocrinol Metab. 1997;82:1894–1898. - PubMed
-
- Abu Shehab M., Khosravi J., Han V.K., Shilton B.H., Gupta M.B. Site-specific IGFBP-1 hyper-phosphorylation in fetal growth restriction: clinical and functional relevance. J Proteome Res. 2010;9:1873–1881. - PubMed
-
- Nissum M., Abu Shehab M., Sukop U., Khosravi J.M., Wildgruber R., Eckerskorn C., Han V.K., Gupta M.B. Functional and complementary phosphorylation state attributes of human insulin-like growth factor-binding protein-1 (IGFBP-1) isoforms resolved by free flow electrophoresis. Mol Cell Proteomics. 2009;8:1424–1435. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

