Systematic Analysis of Human Cells Lacking ATG8 Proteins Uncovers Roles for GABARAPs and the CCZ1/MON1 Regulator C18orf8/RMC1 in Macroautophagic and Selective Autophagic Flux
- PMID: 29038162
- PMCID: PMC5730722
- DOI: 10.1128/MCB.00392-17
Systematic Analysis of Human Cells Lacking ATG8 Proteins Uncovers Roles for GABARAPs and the CCZ1/MON1 Regulator C18orf8/RMC1 in Macroautophagic and Selective Autophagic Flux
Abstract
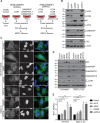
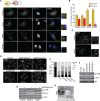
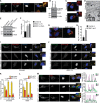
Selective autophagy and macroautophagy sequester specific organelles/substrates or bulk cytoplasm, respectively, inside autophagosomes as cargo for delivery to lysosomes. The mammalian ATG8 orthologues (MAP1LC3A/B/C and GABARAP/L1/L2) are ubiquitin (UB)-like proteins conjugated to the autophagosome membrane and are thought to facilitate cargo receptor recruitment, vesicle maturation, and lysosomal fusion. To elucidate the molecular functions of the ATG8 proteins, we engineered cells lacking genes for each subfamily as well as all six mammalian ATG8s. Loss of GABARAPs alone attenuates autophagic flux basally and in response to macroautophagic or selective autophagic stimuli, including parkin-dependent mitophagy, and cells lacking all ATG8 proteins accumulate cytoplasmic UB aggregates, which are resolved following ectopic expression of individual GABARAPs. Autophagosomes from cells lacking GABARAPs had reduced lysosomal content by quantitative proteomics, consistent with fusion defects, but accumulated regulators of late endosome (LE)/autophagosome maturation. Through interaction proteomics of proteins accumulating in GABARAP/L1/L2-deficient cells, we identified C18orf8/RMC1 as a new subunit of the CCZ1-MON1 RAB7 guanine exchange factor (GEF) that positively regulates RAB7 recruitment to LE/autophagosomes. This work defines unique roles for GABARAP and LC3 subfamilies in macroautophagy and selective autophagy and demonstrates how analysis of autophagic machinery in the absence of flux can identify new regulatory circuits.
Keywords: ATG8; autophagosome maturation; macroautophagy; mitophagy; selective autophagy.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
A new perspective on the autophagic and non-autophagic functions of the GABARAP protein family: a potential therapeutic target for human diseases.Mol Cell Biochem. 2024 Jun;479(6):1415-1441. doi: 10.1007/s11010-023-04800-5. Epub 2023 Jul 13. Mol Cell Biochem. 2024. PMID: 37440122 Review.
-
Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation.J Cell Biol. 2016 Dec 19;215(6):857-874. doi: 10.1083/jcb.201607039. Epub 2016 Nov 18. J Cell Biol. 2016. PMID: 27864321 Free PMC article.
-
Autophagosome formation and cargo sequestration in the absence of LC3/GABARAPs.Autophagy. 2017 Apr 3;13(4):772-774. doi: 10.1080/15548627.2017.1281492. Epub 2017 Feb 6. Autophagy. 2017. PMID: 28165849 Free PMC article.
-
Methods for Studying Interactions Between Atg8/LC3/GABARAP and LIR-Containing Proteins.Methods Enzymol. 2017;587:143-169. doi: 10.1016/bs.mie.2016.10.023. Epub 2017 Jan 12. Methods Enzymol. 2017. PMID: 28253953
-
LC3/GABARAP family proteins: autophagy-(un)related functions.FASEB J. 2016 Dec;30(12):3961-3978. doi: 10.1096/fj.201600698R. Epub 2016 Sep 6. FASEB J. 2016. PMID: 27601442 Review.
Cited by
-
PINK1-dependent mitophagy is driven by the UPS and can occur independently of LC3 conversion.Cell Death Differ. 2019 Aug;26(8):1428-1441. doi: 10.1038/s41418-018-0219-z. Epub 2018 Oct 30. Cell Death Differ. 2019. PMID: 30375512 Free PMC article.
-
The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress.J Biol Chem. 2019 May 17;294(20):8197-8217. doi: 10.1074/jbc.RA118.002829. Epub 2019 Mar 29. J Biol Chem. 2019. PMID: 30926605 Free PMC article.
-
A new perspective on the autophagic and non-autophagic functions of the GABARAP protein family: a potential therapeutic target for human diseases.Mol Cell Biochem. 2024 Jun;479(6):1415-1441. doi: 10.1007/s11010-023-04800-5. Epub 2023 Jul 13. Mol Cell Biochem. 2024. PMID: 37440122 Review.
-
Autophagosome maturation: An epic journey from the ER to lysosomes.J Cell Biol. 2019 Mar 4;218(3):757-770. doi: 10.1083/jcb.201810099. Epub 2018 Dec 21. J Cell Biol. 2019. PMID: 30578282 Free PMC article. Review.
-
The functions of Atg8-family proteins in autophagy and cancer: linked or unrelated?Autophagy. 2021 Mar;17(3):599-611. doi: 10.1080/15548627.2020.1749367. Epub 2020 Apr 19. Autophagy. 2021. PMID: 32255730 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
